?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Skin cancer is a prevalent type of cancer that affects millions of people globally. However, detecting it can be a challenging task, even for specialized dermatologists. Early detection is crucial for successful treatment, and deep learning techniques, particularly deep convolutional neural networks (DCNNs), have shown tremendous potential in this area. However, achieving high accuracy results requires large volumes of data for training these DCNNs. Since medical organizations and institutions, individually, do not usually have such amounts of information available, and due to the current regulations regarding intellectual property and privacy of medical patient data, it is difficult to share data in a direct way. The primary objective of this work is to overcome this issue through a federated learning approach. We created a privacy-preserving and accurate skin cancer classification system that can assist dermatologists and specialists in making informed patient care decisions. The federated learning DCNNs architecture uses a combination of convolutional and pooling layers to extract relevant features from skin lesion images. It also includes a fully connected layer for classification. To evaluate the proposed architecture, we tested it on three datasets of varying complexity and size. The results demonstrate the applicability of the proposed solution and its efficiency for skin cancer classification.
Introduction
Skin cancer is one of the most common types of cancer worldwide, and its incidence has been increasing over the past few decades (Abarca and Chávez Citation2023). According to the World Health Organization (WHO), there are around 3 million cases of non-melanoma skin cancer and more than 132 cases of melanoma skin cancer diagnosed each year globally (Organization Citation2017). Moreover, skin cancer is responsible for a significant number of deaths every year, with approximately 66,000 deaths attributed to melanoma skin cancer in 2020 alone (Parker Citation2021).
The incidence of skin cancer varies widely across different regions and populations, with higher rates observed in countries with fair-skinned populations and high levels of ultraviolet radiation exposure (Vaccarella et al. Citation2023). For example, Australia has the highest incidence of skin cancer in the world, with two to three times higher rates of melanoma skin cancer than the United States, Europe, and Canada. Similarly, New Zealand has the highest incidence of melanoma skin cancer globally (Venugopal et al. Citation2023).
Accordingly, early detection and accurate diagnosis of skin cancer are critical for improving patient outcomes and reducing mortality rates. However, diagnosis of skin cancer can be challenging (Bibi et al. Citation2023), and visual inspection by dermatologists can be subjective and time-consuming (Dobre et al. Citation2023).
Machine learning, specifically DCNNs, has shown promise in accurately classifying skin lesions in medical images (Gouda et al. Citation2022). Many research works demonstrate the ongoing research efforts in the field of skin cancer classification using machine learning approaches and highlight the potential of these techniques to improve early detection and diagnosis of skin cancer (Dillshad et al. Citation2023). However, these models require large volumes of data for their learning, an infrequent characteristic in real contexts, since medical organizations and institutions, individually, do not usually have such amounts of information available. Moreover, there is still a need for further research to improve the accuracy and reliability of skin cancer classification using machine learning. One of the challenges in this research is to develop an efficient and effective DCNNs architecture that can accurately classify skin lesions using medical images (Abdou Citation2022). It is important to explore the use of different datasets to train and test the DCNNs models to ensure that they are robust and generalizable across different populations and regions.
Federated Learning is a machine learning method that allows models to gain experience from different data sets located in different places without sharing the training data (Wang et al. Citation2021). This is used to train other machine learning algorithms using multiple local data sets without exchanging data, allowing healthcare parties to create a shared global model without putting the training data in a central location. This is very useful, for example, in the case of skin cancer, where hospitals cannot share data due to the privacy of patients. Implementing federated learning in real-world medical workflows, however, presents several challenges. Data heterogeneity, communication overheads, and the balance between model complexity and computational efficiency are notable concerns. Additionally, maintaining data privacy and security, adhering to regulatory standards, and ensuring the model’s integration into clinical practices require meticulous attention.
In this work, our ultimate goal is to develop a privacy-preserving and accurate skin cancer classification system that can assist dermatologists in making informed decisions about patient care. Generally, the main contribution of this work is the development of a federated learning approach to classify skin cancer as malign or benign through the application of convolutional deep learning model. We tested the proposed architecture on three datasets to demonstrate its efficiency.
The remainder of this paper is organized as follows: Section II provides a discussion on the related work. Section III elaborates on the proposed DCNNs architecture. Experimental results were described in Section IV. V. Federated model experimentations have been conducted in Section V, and finally, we concluded our work in Section VI.
Related Work
The field of skin cancer classification using machine learning techniques has seen significant growth in recent years, driven by the need for accurate and efficient diagnosis of skin cancer (Sm, Aravindan, and Appavu Citation2023). Various CNNs have been applied to medical images of skin lesions, achieving promising results. In this section, we review some of the recent studies that have addressed the challenge of skin cancer classification using machine learning approaches.
Karri, Annavarapu, and Acharya (Citation2023) proposed a transfer learning-based approach with an attention mechanism for skin lesion classification. The authors used publicly available datasets and achieved state-of-the-art performance with an accuracy of 97.3%. The proposed approach can potentially improve the reliability and accuracy of skin cancer diagnosis. However, the reliance on publicly available datasets may not fully represent the diversity of skin lesions seen in clinical settings, potentially limiting the model’s generalizability to real-world applications.
Aloui et al. (Sinha and Gupta Citation2022) compares the performance of different deep learning techniques for skin lesion classification, including Convolutional Neural Networks (CNNs), Residual Networks (ResNets), and Dense Convolutional Networks (DenseNets). The authors used the ISIC 2017 dataset and achieved an accuracy of 91.6% with the DenseNet model. The study highlights the importance of selecting appropriate deep learning architectures for skin lesion classification. While promising, the study underscores the challenge of selecting the most effective architecture, suggesting that there is no one-size-fits-all solution and that the performance can vary significantly depending on the dataset’s characteristics.
K. Singh et al. (Khamparia et al. Citation2021) proposes an Inception-v3 based deep learning model for skin lesion classification using dermoscopy images. The authors used the ISIC 2018 dataset and achieved an accuracy of 91.26%. The proposed model highlights the potential of specific architectures but also points to a common limitation in deep learning models: the requirement for substantial computational resources, which can be a barrier for deployment in resource-constrained environments.
In Dutta, Kamrul Hasan, and Ahmad (Citation2021), Sahoo et al., conducted a novel multimodal approach for skin lesion classification using both dermoscopy and clinical images. The authors used a combination of CNN architectures and achieved an accuracy of 89.5%. The proposed approach can potentially improve the accuracy and reliability of skin cancer diagnosis. However, the integration of multimodal data introduces complexity in data preprocessing and model training, presenting a challenge in achieving consistent performance across different image types and sources.
Nawaz et al. (Milton Citation2019) proposes an ensemble of deep CNNs for automated skin lesion classification. The authors used the ISIC 2019 dataset and achieved an accuracy of 90.39%. The proposed approach can potentially improve the accuracy and reliability of skin cancer diagnosis. Yet, ensemble methods can suffer from increased model complexity and interpretability issues, making it difficult for clinicians to understand the basis for a model’s predictions.
Kumar et al. (Varma et al. Citation2022) conducted a deep learning-based ensemble model for skin lesion classification using dermoscopy images. The authors used the ISIC 2018 dataset and achieved an accuracy of 91.75%. However, these studies collectively highlight several limitations inherent in the current state-of-the-art, including the dependence on high-quality, annotated datasets, the challenge of model generalization across diverse patient populations, and the computational demands of sophisticated models.
In advancing skin lesion classification, Moldovanu et al. (Citation2023) enhanced the Simple Linear Iterative Clustering (iSLIC) algorithm to utilize superpixels effectively in dermoscopy images. Their approach segments and extracts critical geometric and shape features from skin lesions, avoiding false negatives. The extracted features are analyzed using a variety of machine learning models, including ensemble methods, classical algorithms, and neural networks. Their method demonstrated improved accuracy over existing methods on the 7-Point MED-NODE and PAD-UFES-20 datasets, highlighting the efficacy of combining advanced segmentation techniques with diverse analytical models in dermatological diagnostics.
In their exploration of skin lesion classification, Moldovanu et al. (Citation2021) introduced a novel diagnostic aid that combines surface fractal dimensions and color cluster features, analyzed through k-nearest neighbor with 5-fold cross validation and a Radial basis function neural network (RBFNN). They uniquely apply a 2D Higuchi fractal dimension method for computing surface complexity and employ a clustering method to assess relevant color distributions within lesions. Their findings, tested on the 7-Point, Med-Node, and PH2 databases, reveal that the RBFNN model significantly outperforms the kNN algorithm in accuracy, demonstrating the potential of integrating fractal and color features for precise skin cancer classification.
summarizes some of the strengths and weaknesses of the discussed relevant works. Overall, these publications demonstrate the ongoing research efforts in the field of skin cancer classification using CNNs approaches and highlight the potential of these techniques to improve early detection and diagnosis of skin cancer.
Table 1. Overview of Strengths and Weaknesses of Key Related works on Machine Learning Techniques for Skin Lesion Classification.
Here is a comparative table summarizing the strengths and weaknesses of the discussed papers on skin lesion classification using machine learning techniques:
Proposed Architecture
In this section, we explain the proposed federated learning approach and the convolutional deep learning model. We give more details on federated leaning strategy and the data augmentation, parameter definition, and training method.
Federated Learning Approach
shows the overall architecture of the proposed federated learning approach for skin cancer detection using DCNNs model, where you can see a federated system in which different hospitals (clients) exchange the results of learning with a central server that will act as a data aggregator to finally share this combination back to the local nodes.
Figure 1. Federated learning architecture sample, where a number of clients create local models. The knowledge of each one is transmitted securely to a central server and this combines the information. The resulting model is returned to the clients.
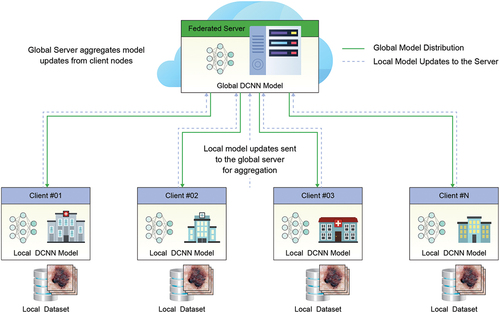
In each hospital or client, the DCNNs model will be deployed on the skin cancer data and will be trained in a totally independent way. The data in each hospital will be divided based on the initial data of some complete datasets that will be presented in the following sections, trying to simulate different situations where a hospital has huge data, other situations where the data is less, and the distribution is not balanced.
All the trained models will be evaluated with three validation data sets (explained in sub-section IV-B). In this way, they can be compared among themselves given that all models will always be evaluated with these sets and have more or less data for training. It should be noted that the federated models will all be trained based on the Federated Average strategy (FedAvg) (McMahan et al. Citation2017). In a simplified way, each client will train its own local model with its own data and each specific number of turns of the dataset, the pairs will be sent. Parameters of the models to the global server that will combine them. This process will be carried out iteratively until the complete training is completed.
FedAvg is a typical implementation of the federated optimization algorithms (Li et al. Citation2020) with a fraction of customers C = 1 and a fixed learning rate η, which makes each client k compute gk = Δ Fk (wt), the mean gradient of the local data in the current model wt, and the core server adds these gradients and applies the update as follows Equation:
Accordingly, each client performs gradient descent locally on the current model using its local data, and the server takes a weighted average of the resulting models.
Convolutional Deep Learning Model
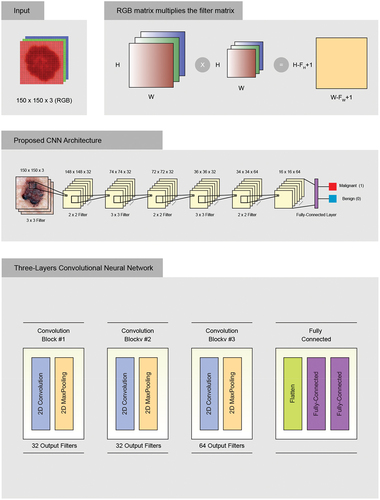
elaborates the convolutional deep learning model adapted from (Carbonell and Peña Citation2021). DCNNs image classifications take an input image, process it, and classify it based on its class criteria. Networks view an image as a matrix of pixels, and the dimensions depend on the resolution of the image. The dimensions of an image are represented as three dimensions which are (H, W, and D). H represents the height, W represents the width, and D is the dimension. In this work, the images are based on the RGB color scheme, where the input images will be (150×150× 3), and the number (3) represents to the values of RGB.
Considering the convolution, the values of the (150×150) input image were plugged in which the image pixel values are (0 and 1) and a (3×3) filter matrix as shown in . Different filters have been applied to convolution of the images to find edge sharpening, detection, and blur. The value of displacement is one, which means the filter will advance one by one at each time.
Next, we had a (148×148) matrix. The depth filter is 32. The ReLu function was applied as an activation function. The ReLu function guarantees a higher performance over tanh or sigmoid in case of image classification problems. The main goal of ReLu function is to minimize the nonlinearity of the DCNNs.
The next step is to minimize the parameter count through the Max Pooling grouping method. The Max Pooling grouping method selects the most important characteristic among all the other features. Later, our matrix will look like this after applying a Max Pool of (2×2) and a stride of 2 by default.
Finally, we end with adding convolutional layers and pooling layers until the DCNNs extract the most important characteristics. We stop adding more convolutions until it begins to extract the low important characteristics. Finally, we flatten our matrix into a vector and transfer it into a fully connected layer that represents the neural network using the Backpropagation algorithm as shown in .
shows that 64 neurons will enter as parameters, forming a vector, and finally, it will give us other output parameters. We define it as 64 because flattening gives us these many parameters. We carry out this process once more, but on a single neuron, which helps us extract only one response. In order to classify the image as malignant or benign cancer, we applied the sigmoid activation function.
This proposed network was designed to achieve better performance on the set of dataset images. A sequential model was used, where the input layer is based on RGB type, and the images pass through three convolutional blocks. Small (3 by 3) filters are used, and each block includes a 2D convolution operation, which switches between blocks. All hidden layers are equipped with a ReLu as the activation function layer, which stands for non-linearity operation, and include spatial pooling by using a maximum pooling layer. The network ends with a classifier block consisting of one layer. The final fully connected output layer performs binary classification, and the activation function is sigmoid.
Data Augmentation
To maximize the use of our limited training examples and improve the model’s accuracy, various random transformations were applied to the data (Maharana, Mondal, and Nemade Citation2022). These included size rescaling, 255-degree rotation, horizontal scrolling, and image zoom. Data augmentation is also expected to prevent overfitting, which is a common issue in machine learning when the model, exposed to very few examples, learns patterns that are not generalized to new data. This enhances the model’s ability to generalize. Additionally, data augmentation helps to balance the dataset by generating the same number of images for each class, enabling a fair comparison of results.
Parameter Definition
To achieve the best performance of the proposed network in relation to the problem, certain parameters must be considered during training. These parameters are:
Batch size: This parameter determines the number of training images processed in forward or backward passes. It is important to note that larger batch sizes require more memory. Our experiments utilized a batch size of 32.
Iterations: The number of iterations refers to the number of forward or backward passes, with each step using a certain number of images equal to the batch size.
Epochs: The number of epochs measures how many times each image has been seen during training. In other words, one epoch means that each image has been seen once, or that there has been a complete forward and backward pass of all the training examples. We used 100 epochs in our experiments.
Loss function: Evaluates the penalty between the prediction and the labeling of the fundamental truth in each batch.
Learning rate: This parameter defines the step size for updating the weights of a model with respect to stochastic gradient descent. The learning rate used in our experiments is 0.001.
Optimizer: There are many optimizers to find the optimal set of parameters for the model. Examples of optimizers include SGD, RMSprop, and ADAM. We used ADAM optimizer in our experiments.
Training Method
The proposed training methodology is conducted to improve the solution of our purposed model. The training data is first processed by the learning method selected by each model, which utilizes stochastic gradient descent. Once the model has learned the weights, a prediction algorithm is used to classify the validation data based on the training data. To evaluate the model’s performance, the predictions are compared with the ground truth data.
Experimental Results
In this section, we describe the dataset and evaluation metrics used in this study. Additionally, we discuss the experimental results of the proposed architecture.
Datasets
We tested the proposed approach for skin cancer detection on three datasets. A brief description on each dataset is given in the following paragraphs.
ISIC 2018 Dataset
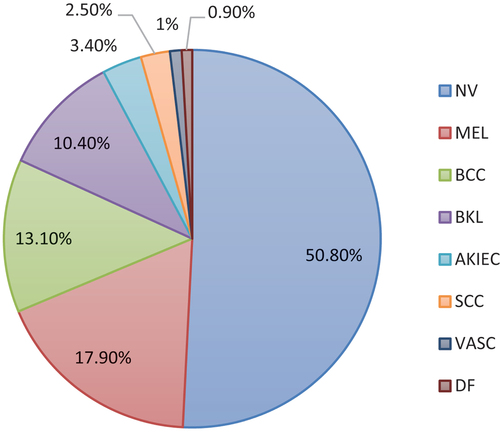
The aim of skin lesion analysis toward melanoma detection (Ali et al. Citation2019) is to detect melanoma using three tasks: Lesion Segmentation, Lesion Attribute Detection, and Disease Classification. The dataset used for training includes 10,015 skin lesion images from seven types of skin diseases, which are Melanoma, Melanocytic nevus, Basal cell carcinoma, Actinic keratosis, Benign keratosis, Dermatofibroma, and Vascular. Melanoma has the highest number of images (1113), followed by Melanocytic nevus (6705), Basal cell carcinoma (514), Actinic keratosis (327), Benign keratosis (1099), Dermatofibroma (115), and Vascular (142). The validation dataset comprises of 193 images. Some examples of these eight types of skin lesions are shown in . The objective of the third task, Disease Classification, is to improve automated predictions of disease classification in dermoscopic images. The possible disease categories are shown below.
PH2 Dataset
The dermoscopic images utilized in this dataset (Mendonça et al. Citation2015) were procured from the Dermatology Service of Hospital Pedro Hispano in Matosinhos, Portugal. The images were captured using a Tuebinger Mole Analyzer system under standardized conditions, with a magnification of 20 × . These images are 8-bit RGB color images with a resolution of 768 × 560 pixels. The PH2 database used in this research contains a total of 200 dermoscopic images of melanocytic lesions, comprising 80 common nevi, 80 atypical nevi, and 40 melanomas. Each image in the database has been medically annotated, including medical segmentation of the lesion, clinical and histological diagnosis, and the evaluation of various dermoscopic criteria such as colors, pigment network, dots/globules, streaks, regression areas, and blue-whitish veil. The assessment of each parameter was performed by an expert dermatologist, according to predefined criteria.
According to the classification process parameters, a hundred epochs were chosen based on the behavior examination of the precision/loss graphs versus the epochs count, where each epoch spent 300 s of the utilized GPU. We then performed a tuning method with a very small learning rate. Additionally, we adapted the Adam optimizer (Zhang Citation2018) to positively minimizing the loss function.
The image counts for training and testing in our experiments are as follows:
ISIC 2018 Dataset: Training: 8,012 images; Test: 2,003 images
PH2 Dataset: Training: 160 images; Test: 40 images
Combined Dataset: Training: 8,172 images; Test: 2,043 images
Evaluation Metrics
In the context of evaluating classification models, various metrics can be used to assess the overall performance. The confusion matrix is a table that displays the counts of the actual versus predicted classifications provided by the model. From this matrix, several key metrics are typically calculated:
Accuracy: Calculates the number of correct predictions divided by the total number of samples.
Sensitivity: Calculates the proportion of true positives. As the sensitivity approaches 100%, the more likely a patient is to have a disease.
Precision: Calculates the fraction of relevant instances among the retrieved instances.
Specificity: Calculates the proportion of time in which a result gives true negatives. As the specificity approaches 100%, it is more likely that the result means that the patient does not have a disease.
Training
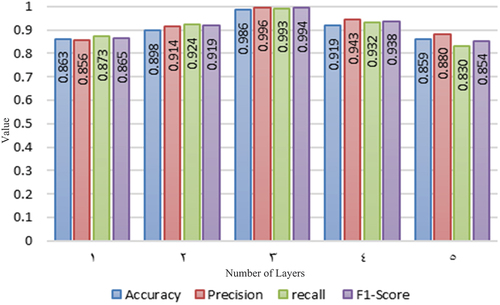
To achieve the best results during the training process, a convolutional layer was added on top of the layer with the highest accuracy. The accuracy values for the PH2 dataset, which contains the least number of images and allows for faster training to determine the best architecture, are shown in .
The F1-Score metric aims to achieve a balance between precision and recall. In the table, we can observe that the F1-Score for three layers indicates good precision and recall. Therefore, we can proceed with training using the architecture with three layers. The following results in are presented for each dataset based on the metrics obtained from the tests carried out on three layers.
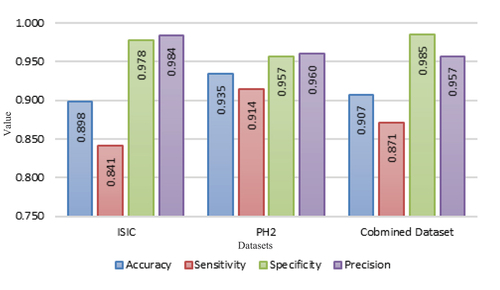
The results were compared for each dataset separately and also for the combined dataset, as shown in . The objective of this comparison is to evaluate the performance of each dataset individually, instead of determining which one is superior, due to the limited number of images in the PH2 dataset.
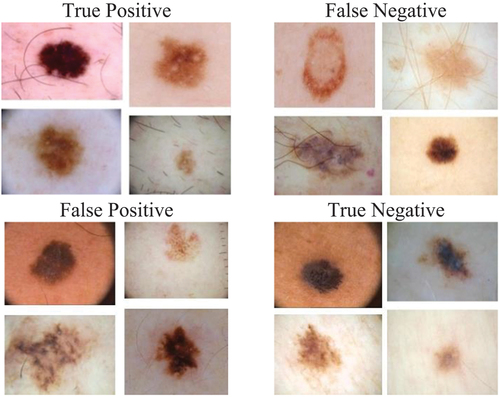
displays the training images used to diagnose malignant or benign cancer. It highlights how the neural network’s errors can impact the similarity or clarity of the image. This serves as an example of how the convolutional neural network processes the input and produces the final diagnosis of benign or malignant, without revealing the internal details of the process. This is because neural networks perform the parameter adjustments and processing internally.
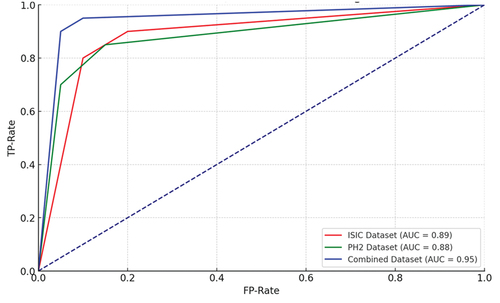
Finally, shows the ROC curve, which is computed to evaluate the performance of the binary classification. In all the datasets, ISIC, PH,2 and combined dataset, the ROC curve depicts the sensitivity versus specificity for classifying malignant and benign cancer. The proposed approach yielded a higher number of true positives than false positives, indicating that the discrimination threshold can accurately classify dermoscopic images, specifically on the PH2 dataset.
Validation
The results of the proposed model demonstrate that good performance can be achieved by utilizing the originally proposed architecture for image classification datasets. This suggests that the proposed model is able to generalize well to a diverse range of classification problems, even though the input images were not included in the training dataset. represents a summary of our experimental results.
Table 2. Experimental results.
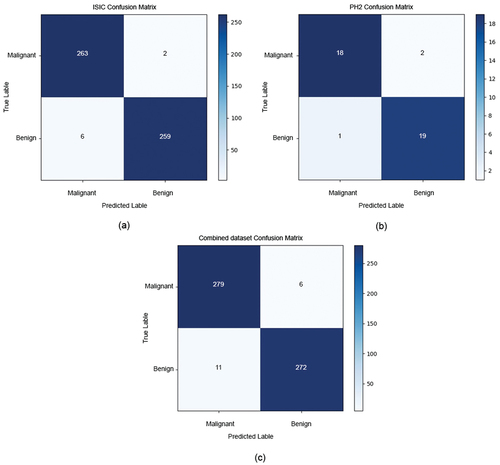
The confusion matrix, which depicts the model’s classification performance on the validation dataset, is presented in . The evaluation of the model should consider the number of true melanomas classified as benign cases. It is important to note that this scenario represents the worst-case scenario, where the model fails to detect a real case of melanoma, which could result in a dangerous and potentially life-threatening situation for the patient.
displays the confusion matrix of the three datasets. shows 265 malignant and 265 benign images for the ISIC dataset. Out of these, 263 were correctly predicted, while 2 were classified as benign. For benign images, 6 were classified as malignant, and 259 were correctly predicted as benign.
shows the confusion matrix of the PH2 dataset, which contains 20 malignant and 20 benign images, where 18 were correctly predicted, and 2 were classified as benign.
For benign images, 1 was classified as malignant, and 19 were correctly predicted as benign. shows the confusion matrix of the combined dataset, which contains 285 malignant and 285 benign images. Out of these, 279 were correctly predicted, while 6 were classified as benign. For benign images, 11 were classified as malignant, and 272 were correctly predicted as benign.
Federated Model Experimentations
Given the existing problems in the transmission, treatment and privacy of skin cancer data, this work conducted simulation experiments on the use of a federated model with data distributed in different institutions such as hospitals compared to a local model. More precisely, four experiments have been carried out both in centralized and federated architectures. We perform the experiments as follows:
First experiment: A single client with a local model trained on 100% of the training data.
Second experiment: Two clients, a local model for each one with the distribution of 40% and 60% of the training data in each one.
Third experiment: Federated model of the two clients with the aforementioned data distributions.
Fourth experiment: Federated model of a single global server and five clients with local models with training dataset distributions of 15%, 15%, 20%, 20%, and 30% of the data.
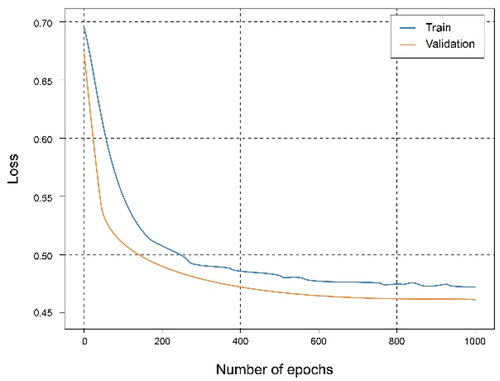
In these experiments, different types of parameters and metrics have been used for the evaluation of the experiments, such as accuracy, loss, and the confusion matrix. A total of 1000 epochs have been established, and accordingly, the dataset will iterate thousand times while training is conducted. The evaluation is carried out each season with the test dataset, while the federated approach established 100 training rounds and 10 epochs. Then, in the federated architecture, local models are combined and updated on the network. Server aggregation every 10 epochs of local training and the evaluation is performed after those 10 epochs. Finally, we analyzed how it affects having a higher or lower volume of data in the clients and if the federation in different cases helps to improve the results.
First Experiment
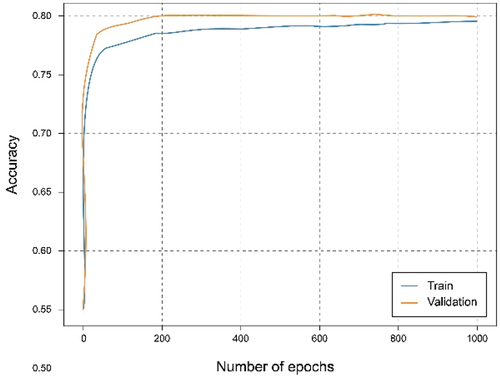
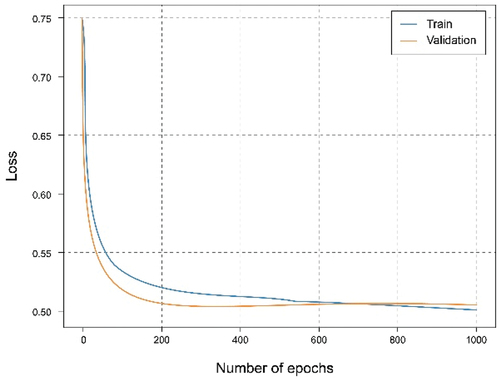
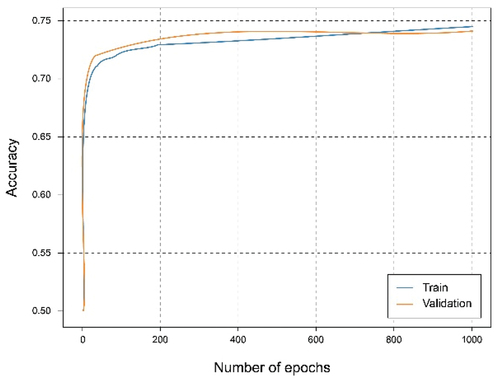
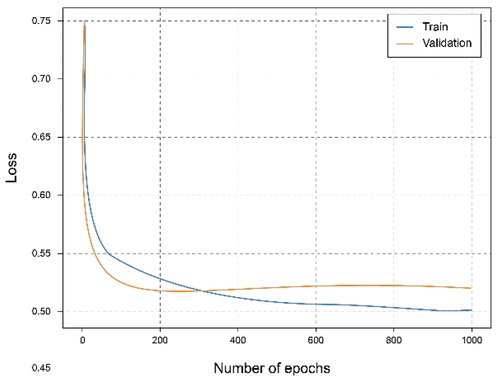
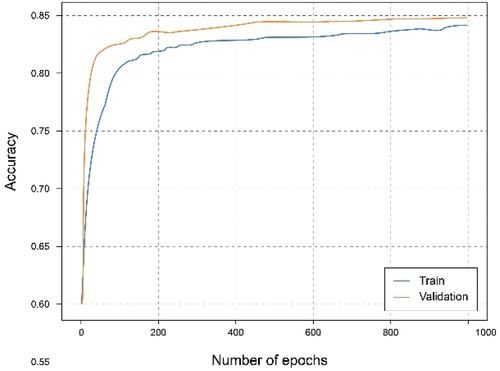
shows that using the overall dataset obtained a good. It can be observed that the Accuracy graph shows that the model learns and predicts with 81% probability of success. represents the loss graph, and it can be seen that a reduced error value is reached, declining during training, without noticing symptoms of overfitting or underfitting.
Second Experiment
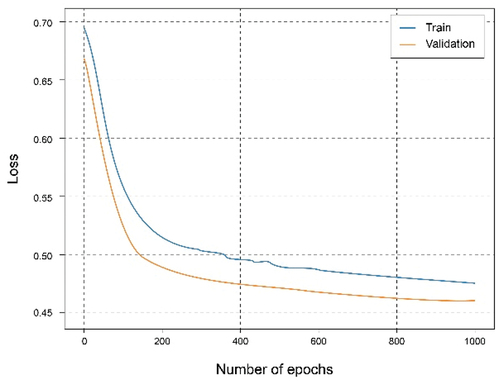
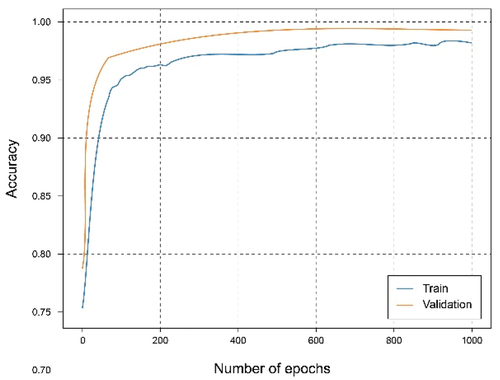
In this experiment, the dataset has been split for two clients in which the first client has 40% of the training data and the second client has 60%. The training has been carried out for both clients. It can be seen from the accuracy graph in that from epoch number 400, a certain overfitting is observed for the model of the first client. This is because the number of training data has been reduced and the model, having a complex architecture, where it learns too much from the training data, so it is not capable of generalizing in the test set. This is corroborated in where the loss starts to grow instead of shrinking. In this case, the metrics are a little worse than in the case of the client with the complete dataset since this overfitting has occurred, which has not been too high, allowing the model to continue and work smoothly.
In the case of the second client, the overfitting problems are not so exaggeratedly appreciated, since the data size is larger. Analyzing both , we can observe a similar result compared to those of the client with the full dataset as there is a greater volume of data.
Third Experiment
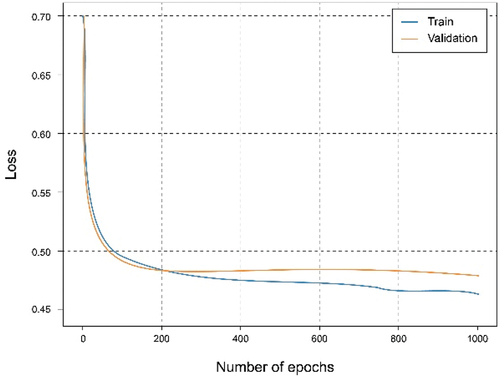
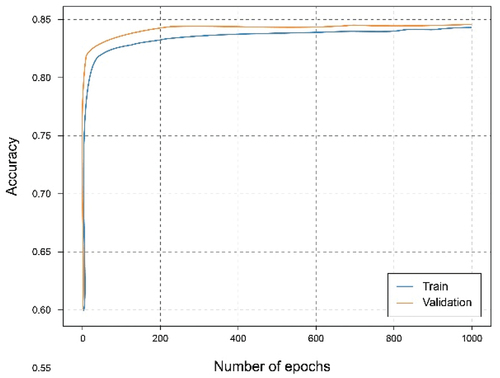
In this experiment, a federated architecture consisting of two clients and one global server has been simulated. The clients, with a distribution of the training set of 40% and 60%. Observing , it can be seen that they reach almost the same results as when using the centralized architecture with 100% of the data and training. It is also observed an interesting phenomenon and that is that the curves relative to the set of tests exceed those of training, which means that the learning is being carried out in an adequate and no observation of overfitting or underfitting, as shown in .
Fourth Experiment
In this experiment, five client’s architecture has been created. The distribution of the training data set is divided as follows: two clients with 15%, two with 20% and the reaming client has 30% of the training dataset. They will all train independently to observe the results. The goal here is to simulate the case where certain clients (large hospitals) have a lot of data and other clients (local health centers) have less data.
Analyzing , it can be observed that by combining the local models through a federated strategy on the global server, the results improved, obtaining an accuracy approximate 95% and a fairly small loss value. Something similar happens in the federated case of two clients where each one contributes different knowledge that combined allows generalize better and make more accurate predictions. It should be noted that in this case, there are clients nodes with less data, therefore the result comes to resemble or even exceed that of the local client with all the starting data.
Discussion
In this work, we proposed an effective approach for skin cancer classification using deep convolutional neural networks (DCNNs) through a federated learning approach. By evaluating our proposed architecture on three distinct datasets, we achieved commendable classification accuracy, demonstrating the potential of federated learning in enhancing privacy-preserving, accurate medical diagnosis systems. The utilization of federated learning addresses critical challenges associated with data privacy and accessibility, showcasing a scalable model for collaborative, decentralized machine learning without compromising patient data.
Our findings indicate a promising direction, with accuracy rates competitive with state-of-the-art methods in skin cancer classification, as shown in . For instance, Karri, Annavarapu et al.'s approach (Karri, Annavarapu, and Acharya Citation2023) yielded a 97.3% accuracy using a transfer learning-based method with an attention mechanism. While their method reported higher accuracy, our federated learning approach introduces an additional layer of data privacy and security, addressing significant concerns in medical data handling. Similarly, Aloui et al. (Sinha and Gupta Citation2022) achieved a 91.6% accuracy with DenseNet models on the ISIC 2017 dataset. Our federated model, particularly in the fourth experiment involving multiple clients, showed a nearly 95% accuracy, highlighting the efficacy of federated learning in leveraging data from diverse sources to improve model performance.
Table 3. Comparison with related works.
The novelty of our approach lies in its ability to maintain high accuracy while ensuring data privacy, a crucial aspect often overlooked in centralized machine learning paradigms. Additionally, the flexibility and scalability of federated learning, as demonstrated through various experimentations, suggest its applicability across different institutions and data sizes, a significant advantage over traditional models that require centralized data pooling.
Despite these promising results, our study encounters limitations inherent to federated learning and the specific architecture employed. First, the data heterogeneity across different institutions poses a challenge, potentially leading to model bias or underperformance on certain types of skin lesions not well represented in the training datasets. This is a common issue in machine learning models reliant on diverse data sources, emphasizing the need for more inclusive and comprehensive data collection strategies.
Second, the computational and communication overhead associated with federated learning, especially in scenarios involving numerous clients with varying data sizes, can affect efficiency. Optimizing model training and update aggregation processes is critical to mitigating these issues, ensuring the model’s scalability and practicality in real-world applications.
Third, while federated learning enhances data privacy, it does not eliminate all privacy and security concerns. Potential vulnerabilities could still be exploited through model inversion attacks or inference from aggregated updates, necessitating ongoing research into more robust privacy-preserving techniques.
Lastly, the integration of our federated learning model into clinical workflows remains a challenge. The practical deployment requires not only technological compatibility but also the acceptance and trust of medical professionals. Further studies focusing on user-friendly interfaces, interpretability of model decisions, and validation in clinical settings are essential to bridging the gap between technical innovation and practical application.
Conclusion
The global prevalence of skin cancer underscores the critical need for advancements in early detection methods, which are paramount for effective treatment outcomes. This study leverages artificial intelligence, specifically deep convolutional neural networks (DCNNs), to address the challenge of skin cancer classification. Our work not only showcases the potential of DCNNs in enhancing diagnostic accuracy but also introduces a federated learning approach as a pivotal solution to data privacy and accessibility issues prevalent in medical research.
Our experiments demonstrate the model’s robust performance across various datasets. Notably, for the ISIC dataset, our model achieved a high degree of accuracy, correctly classifying 263 out of 265 malignant images and 259 out of 265 benign images. Similar high accuracy levels were observed with the PH2 dataset and a combined dataset, underscoring the model’s effectiveness in skin cancer detection. Moreover, federated learning experimentations revealed an encouraging model accuracy of up to 98% in scenarios involving multiple clients, emphasizing the feasibility of employing advanced AI techniques in a privacy-preserving manner across distributed data sources.
By demonstrating the effectiveness of federated learning for skin cancer detection, our study contributes significantly to the broader field of medical imaging and AI applications. It highlights the feasibility of employing advanced AI techniques in a privacy-preserving manner, thereby paving the way for future research and practical implementations that could revolutionize healthcare delivery. This approach not only augments the accuracy and efficiency of skin cancer detection but also serves as a model for tackling similar challenges across various domains of medical imaging. The implications of our findings extend beyond skin cancer diagnosis, offering insights into the potential of federated learning and DCNNs to enhance the quality and accessibility of healthcare. As we move forward, focusing on improving the generalizability of our architecture across larger and more diverse datasets, integrating these technologies into clinical workflows, and enhancing the interpretability of AI-extracted features will be crucial. These efforts will not only bolster the effectiveness of diagnostic tools but also contribute to the development of AI-driven solutions that are both innovative and ethically responsible, ultimately leading to better patient care and outcomes. Our study reaffirms the significance of integrating AI in medical diagnostics and sets a foundation for future exploration that could significantly impact the broader landscape of medical imaging and artificial intelligence applications.
Acknowledgements
This work was supported by Research Supporting Project Number (RSPD2024R585), King Saud University, Riyadh, Saudi Arabia.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abarca, J. M. H., and A. J. P. Chávez. 2023. Malignant Nail Melanoma in a case report. Journal of Pharmaceutical Negative Results 14 (2):67–27.
- Abdou, M. 2022. Literature review: Efficient deep neural networks techniques for medical image analysis. Neural Computing and Applications 34 (8):5791–812. doi:10.1007/s00521-022-06960-9.
- Ali, R., Hardie, R.C., De Silva, M.S. and T.M. Kebede. 2019. Skin lesion segmentation and classification for ISIC 2018 by combining deep CNN and handcrafted features. arXiv Preprint arXiv:05730.
- Bibi, S., M. A. Khan, J. H. Shah, R. Damaševičius, A. Alasiry, M. Marzougui, M. Alhaisoni, and A. Masood. 2023. MSRNet: Multiclass skin lesion recognition using additional residual block based fine-tuned deep models information fusion and best feature selection. Diagnostics 13 (19):3063. doi:10.3390/diagnostics13193063.
- Carbonell, M. C., and R. V. Peña. 2021. Convolutional neural network architecture for skin cancer diagnosis. European Journal of Molecular Clinical Medicine 8 (3):2819–33.
- Dillshad, V., M. A. Khan, M. Nazir, O. Saidani, N. Alturki, and S. Kadry. 2023. D2LFS2Net: Multi‐class skin lesion diagnosis using deep learning and variance‐controlled Marine Predator optimisation: An application for precision medicine. CAAI Transactions on Intelligence Technology. doi:10.1049/cit2.12267.
- Dobre, E.-G., M. Surcel, C. Constantin, M. A. Ilie, A. Caruntu, C. Caruntu, and M. Neagu. 2023. Skin cancer pathobiology at a glance: A focus on imaging techniques and their potential for improved diagnosis and surveillance in clinical cohorts. International Journal of Molecular Sciences 24 (2):1079. doi:10.3390/ijms24021079.
- Dutta, A., M. Kamrul Hasan, and M. Ahmad. 2021. Skin lesion classification using convolutional neural network for melanoma recognition. Proceedings of International Joint Conference on Advances in Computational Intelligence: IJCACI 2020, Singapore. Springer.
- Gouda, W., N. U. Sama, G. Al-Waakid, M. Humayun, and N. Z. Jhanjhi. 2022. Detection of skin cancer based on skin lesion images using deep learning. In Healthcare, ed. C. Joaquim and G. Danielec, 2–18. Basel, Switzerland: MDPI.
- Karri, M., C. S. R. Annavarapu, and U. R. Acharya. 2023. Skin lesion segmentation using two-phase cross-domain transfer learning framework. Computer Methods Programs in Biomedicine 231:107408. doi:10.1016/j.cmpb.2023.107408.
- Khamparia, A., P. K. Singh, P. Rani, D. Samanta, A. Khanna, and B. Bhushan. 2021. An internet of health things‐driven deep learning framework for detection and classification of skin cancer using transfer learning. Transactions on Emerging Telecommunications Technologies 32 (7):e3963. doi:10.1002/ett.3963.
- Li, T., Sahu, A.K., Zaheer, M., Sanjabi, M., Talwalkar, A. and Smith, V 2020. Federated optimization in heterogeneous networks. Proceedings of Machine Learning Systems 2:429–50.
- Maharana, K., S. Mondal, and B. Nemade. 2022. A review: Data pre-processing and data augmentation techniques. Global Transitions Proceedings 3 (1):91–99.
- McMahan, B., Moore, E., Ramage, D., Hampson, S. and Arcas, B.A. 2017. Communication-efficient learning of deep networks from decentralized data. In Artificial Intelligence and Statistics, 1273–1282. Florida, USA: MLR, JMLR & W&CP.
- Mendonça, T., Ferreira, P.M., Marçal, A.R., Barata, C., Marques, J.S., Rocha, J. and Rozeira, J. 2015. Ph2: A public database for the analysis of dermoscopic images. Dermoscopy Image Analysis 10:419.
- Milton, M. A. A. 2019. Automated skin lesion classification using ensemble of deep neural networks in ISIC 2018: Skin lesion analysis towards melanoma detection challenge. arXiv Preprint arXiv:10802.
- Moldovanu, S., F. A. Damian Michis, K. C. Biswas, A. Culea-Florescu, and L. Moraru. 2021. Skin lesion classification based on surface fractal dimensions and statistical color cluster features using an ensemble of machine learning techniques. Cancers 13 (21):5256. doi:10.3390/cancers13215256.
- Moldovanu, S., M. Miron, C.-G. Rusu, K. C. Biswas, and L. Moraru. 2023. Refining skin lesions classification performance using geometric features of superpixels. Scientific Reports 13 (1):11463. doi:10.1038/s41598-023-38706-5.
- Organization, W. H. 2017. Radiation: Ultraviolet (UV) radiation and skin cancer.
- Parker, E. R. 2021. The influence of climate change on skin cancer incidence–A review of the evidence. International Journal of Women’s Dermatology 7 (1):17–27. doi:10.1016/j.ijwd.2020.07.003.
- Sinha, S., and N. Gupta. 2022. A comparative analysis of transfer learning-based techniques for the classification of Melanocytic Nevi. arXiv Preprint arXiv:10972.
- Sm, J., C. Aravindan, and R. Appavu. 2023. Classification of skin cancer from dermoscopic images using deep neural network architectures. Multimedia Tools Applications 82 (10):15763–78. doi:10.1007/s11042-022-13847-3.
- Vaccarella, S., D. Georges, F. Bray, O. Ginsburg, H. Charvat, P. Martikainen, H. Brønnum-Hansen, P. Deboosere, M. Bopp, M. Leinsalu, et al. 2023. Socioeconomic inequalities in cancer mortality between and within countries in Europe: A population-based study. The Lancet Regional Health - Europe 25. doi:10.1016/j.lanepe.2022.100551.
- Varma, P. B. S., S. Paturu, S. Mishra, B. S. Rao, P. M. Kumar, and N. V. Krishna. 2022. SLDCNet: Skin lesion detection and classification using full resolution convolutional network-based deep learning CNN with transfer learning. Expert Systems 39 (9):e12944. doi:10.1111/exsy.12944.
- Venugopal, K., D. Youlden, L. T. Marvelde, R. Meng, J. Aitken, S. Evans, I. Kostadinov, R. Nolan, H. Thomas, and K. D’Onise. 2023. Twenty years of melanoma in Victoria, Queensland, and South Australia (1997–2016). Cancer Epidemiology 83:102321. doi:10.1016/j.canep.2023.102321.
- Wang, R., J. Xu, Y. Ma, M. Talha, M. S. Al-Rakhami, and A. Ghoneim. 2021. Auxiliary diagnosis of COVID-19 based on 5G-enabled federated learning. IEEE Network 35 (3):14–20. doi:10.1109/MNET.011.2000704.
- Zhang, Z. 2018. Improved Adam optimizer for deep neural networks. 2018 IEEE/ACM 26th international symposium on quality of service (IWQoS), Banff, AB, Canada. Ieee.