Abstract
The prevalence of Lactobacillus iners is currently difficult to determine since it cannot be propagated on conventional Lactobacillus-selective media. Based on comparative analysis of 16S rRNA gene sequences a simple L. iners-specific PCR method was developed and then applied to screen vaginal, fecal, and saliva samples for this species. Twenty-one (53%) of 40 vaginal specimens were positive, whereas fecal (n=9) and saliva (n=16) samples were PCR-negative for L. iners. These findings support previous molecular and culture-based ecology studies that have indicated L. iners may be relatively more prevalent in the microflora of the human vagina than that of the oral or gastrointestinal cavities.
Introduction
Lactobacilli are indigenous to the oral, gastrointestinal, and vaginal cavities and certain strains have been shown to be effective when introduced either orally Citation[1–3] or intravaginally Citation[4–6]. The study of the ecology of Lactobacillus species is important for our greater understanding of the role of these organisms in enhancing health.
Lactobacillus iners was initially isolated from human urine, vaginal discharges, and endometrial and cervical specimens Citation[7]. L. iners differs from other Lactobacillus species in that it cannot be grown on MRS or Rogosa agar Citation[7]. The identification of Lactobacillus species by classical phenotypic methods is difficult, time-consuming, and frequently yields ambiguous results Citation[8], and for L. iners this is complicated by the absence of suitable selective media. An alternative approach is PCR-based identification using species-specific primers designed to variable regions of the 16S rRNA genes Citation[8–10]. To improve our understanding of the ecology of L. iners a simple PCR-based detection methodology resembling the available techniques for other Lactobacillus species is required. Here we report the development, evaluation, and application of just such a technique. Our findings indicate that L. iners is relatively more frequently present in samples from the vagina than from the intestine or oral cavity and support our hypothesis that it may have a significant ecological role within the microflora of the human vagina.
Materials and methods
Bacterial strains
The L. iners type strain CCUG 28746T was obtained from the culture collection, University of Göteborg, Sweden. L. iners strains BVS011, FB077-03, FB088-01, FB094-05b, FB123-CNA-4, PB2003/087-T2-1, and PB2003/195-T1-1 were obtained from Professor Mario Vaneechoutte (Department of Clinical Chemistry, Microbiology & Immunology, Ghent University Hospital, Belgium). Other bacteria used in this study () were freshly obtained vaginal isolates, clinical isolates or strains from the culture collection of Prof. J.R. Tagg (Microbiology and Immunology Department, University of Otago, New Zealand). Lactobacilli were propagated on either Lactobacillus-Rogosa agar (Difco, USA) or blood agar (Columbia agar base, Difco) with 5% v/v human blood) with incubation under anaerobic conditions (85% N2, 10% H2, and 5% CO2) at 37°C for 48–72 h.
Table I. Bacterial strains used in this study.
Vaginal, saliva, and fecal samples
The Otago Ethics Committee approved the study and all participants provided written consent. The vaginal samples (n = 40) were obtained from healthy women attending prenatal clinics. No Gram-stain grading or testing for bacterial vaginosis was performed. High vaginal swabs (TRANSWAB®, England) were obtained from women by a gynecologist, and kept at 4°C until processing within 30 min. The contents of each swab were dispersed in 3 ml brain heart infusion (BHI, Difco) and the resulting suspension was used to obtain total DNA preparations.
For saliva samples, the subjects were asked to provide 1 ml of saliva by spitting into a sterile bottle. The samples were immediately centrifuged at 15 300 g for 5 min, and the pellets (resuspended in 100 µl H2O) were used to obtain total DNA. InstaGene™ matrix (BioRad Laboratories) was used to obtain total representative DNA from the vaginal and oral specimens, according to the manufacturer's instructions.
Fecal samples were obtained from healthy individuals and kept at 4°C until processing. For DNA extractions, 10 mg of fecal material was washed three times by resuspending in 1.0 ml of phosphate-buffered saline and centrifuging at 14 000 g. The washed pellets were then resuspended in 450 µl of extraction buffer (100 mM Tris-HCl, 40 mM EDTA, pH 9.0) and 50 µl of 10% SDS followed by the addition of 300 mg of glass beads (diameter, 0.1 mm) and 500 µl of buffer-saturated phenol, before vigorous vortex mixing for 30 s using a FastPrep FP 120 (Bio 101, Vista, CA, USA) at a power level of 5.0. After centrifugation at 14 000 g for 5 min, the supernatants were collected. Subsequently, phenol-chloroform extractions were performed, and DNA was obtained by isopropanol precipitation. Finally, the DNA was suspended in 1 ml of TE buffer.
DNA extraction from pure cultures
Extraction from pure cultures was performed using the Qiagen DNAeasy tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, using logarithmic growth phase cells in Todd Hewitt broth (THB, Difco).
Development of species-specific primers for L. iners
Twenty-seven 16S rRNA gene sequences of Lactobacillus species (15 L. iners sequences plus 12 from other lactobacilli identified by BLAST to have sequences that were most closely related to the L. iners consensus sequences) were aligned to facilitate the design of L. iners-specific PCR primers. Alignments and the sequence accession numbers are given in . Alignment was achieved by using the Clustal W alignment method DNAMAN (Lynnon Biosoft, Vaudreuil, Canada) with low gap penalties (0.1 gap open, 0.05 gap extension, 30% delay divergent). The 15 L. iners 16S rRNA sequences were all of those that were available in the public databases. Two regions were identified as specific for L. iners (). The PCR primer pair InersFw (5′-GTC TGC CTT GAA GAT CGG-3′) and InersRev (5′-ACA GTT GAT AGG CAT CAT C-3′) were designed to these regions and synthesized (Invitrogen New Zealand Ltd, Auckland, New Zealand). The chosen primers were based upon positions 42–101 and 162–221 of the sequence AY283264.
Table II. Alignment of 16S rRNA sequences.
PCR analysis
Species-specific PCR reactions for L. iners comprised 3 µl DNA, 1 U Hot start Taq DNA polymerase (Eppendorf AG, Hamburg, Germany), 5 µl of 10× buffer (Eppendorf), 1 µl of PCR nucleotide mix (Eppendorf), and 1 µl (4 ng) of each primer (Invitrogen). The volume was made up to 50 µl with sterile MQ water. After a 3 min initial denaturation at 95°C, 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and elongation at 65°C for 1 min were carried out. The primers and conditions for the PCR reactions for eubacterial, Lactobacillus, and L. crispatus rRNA gene sequences have been described previously Citation[10–12] and these were used in this study to confirm the presence of DNA and absence of PCR inhibitors in the samples. lists all the primers used in this study.
Table III. Primers used in this study.
Results
Sensitivity and specificity of the L. iners selective primers
The specificity of the primers for L. iners was evaluated by carrying out PCR using genomic DNA from the various strains of different genera and species listed in . No cross-reactivity was detected with DNA from any other species, including the most closely related lactobacilli (). To determine the sensitivity of the PCR assay, an 18 h THB culture of L. iners was serially diluted in sterile saline and the numbers of colony-forming units (cfu) were assessed by plating on blood agar. Known numbers of L. iners were then added to clinical samples that had been previously found to be PCR-negative for L. iners before DNA extraction. The PCR lower limit of detection was estimated to be 50 cfu, as addition of this number of cells (10 µl of diluted culture) to clinical specimens (50 µl) led to their conversion from PCR-negative to PCR-positive.
PCR analysis of clinical samples
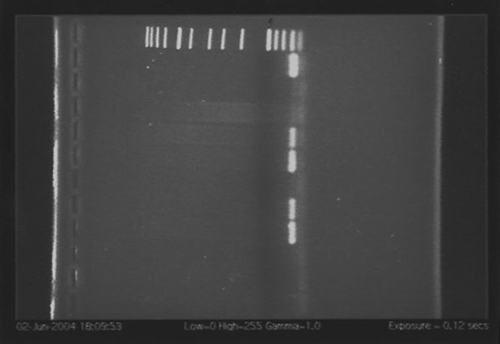
L. iners was detected in 21 (53%) of the 40 tested vaginal specimens, but not in any of the fecal or saliva specimens. shows four of nine representative vaginal specimens exhibiting PCR-positive reactions using the L. iners-specific primers. Amplicons from selected samples were DNA-sequenced and this confirmed the identity of the products to be L. iners. All the samples were PCR-positive for eubacterial DNA. All the vaginal and saliva samples and five of the nine fecal samples were PCR-positive for Lactobacillus DNA. Fourteen of the 40 vaginal samples, one saliva sample, but none of the fecal specimens contained detectable L. crispatus DNA.
Discussion
The acquisition of PCR products for DNA from eubacteria, Lactobacillus, and L. crispatus established that the samples contained amplifiable DNA and also that detection frequencies for other Lactobacillus species were in the normal range for these specimens Citation[7], Citation[13–17]. The relatively frequent detection of L. iners in the vaginal specimens from healthy women indicates that this species could be a significant inhabitant of the normal microbiota of this site. All the tested saliva and fecal samples were PCR-negative for L. iners. Several previous studies Citation[7], Citation[13–17] have also indicated that this species inhabits the vagina, whereas molecular ecological studies of the gastrointestinal Citation[18] and oral ecosystems Citation[19–21] have failed to detected L. iners. The rectum, which is considered to be a primary source of lactobacilli for vaginal microflora maintenance, seems relatively seldom to harbor L. iners (4%) when compared with other Lactobacillus species such as L. crispatus (43%), L. jensenii (23%), and L. gasseri (18%) Citation[22]. On the basis of our present findings using a sensitive and specific PCR-detection method, and previous ecological studies Citation[18–21], it seems that L. iners may indeed have relatively specific affinity for colonization of the human vagina. The primers InersFw and InersRev can be used in many different settings and in different nucleic acid-based ecological applications. The clinical relevance of the changes in vaginal microbiota as it shifts from healthy to dysbiosis (bacterial vaginosis) is not fully understood. L. iners could be associated with early disease, relapse or recovery as either a final protective species or a disease facilitator.
Acknowledgements
Mr Alqumber is supported by a scholarship provided by the Saudi Arabian Ministry of Higher Education.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Fuller R. The importance of Lactobacilli in maintaining normal microbial balance in the crop. Br Poult Sci 1977; 18: 85–94
- Watkins BA, Kratzer FH. Effect of oral dosing of Lactobacillus strains on gut colonization and liver biotin in broiler chicks. Poult Sci 1983; 62: 2088–94
- Watkins BA, Miller BF. Competitive gut exclusion of avian pathogens by Lactobacillus acidophilus in gnotobiotic chicks. Poult Sci 1983; 62: 1772–9
- Reid G, Bruce AW, McGroarty JA, Cheng KJ, Costerton JW. Is there a role for lactobacilli in prevention of urogenital and intestinal infections?. Clin Microbiol Rev 1990; 3: 335–44
- Bruce AW, Reid G. Intravaginal instillation of lactobacilli for prevention of recurrent urinary tract infections. Can J Microbiol 1988; 34: 339–43
- Reid G, Charbonneau D, Erb J, Kochanowski B, Beuerman D, Poehner R, et al. Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: randomized, placebo-controlled trial in 64 healthy women. FEMS Immunol Med Microbiol 2003; 35: 131–4
- Falsen E, Pascual C, Sjoden B, Ohlen M, Collins MD. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol 1999; 49(Pt 1)217–21
- Dickson EM, Riggio MP, Macpherson L. A novel species-specific PCR assay for identifying Lactobacillus fermentum. J Med Microbiol 2005; 54(Pt 3)299–303
- Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005; 353: 1899–911
- Walter J, Tannock GW, Tilsala-Timisjarvi A, Rodtong S, Loach DM, Munro K, et al. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl Environ Microbiol 2000; 66: 297–303
- Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol 1990; 28: 1942–6
- Horie M, Kajikawa HS, Toba T. Identification of Lactobacillus crispatus by polymerase chain reaction targeting S-layer protein gene. Lett Appl Microbiol 2002; 35: 57–61
- Burton JP, Reid G. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J Infect Dis 2002; 186: 1770–80
- Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 2004; 150(Pt 8)2565–73
- Burton JP, Cadieux PA, Reid G. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl Environ Microbiol 2003; 69: 97–101
- Tarnberg M, Jakobsson T, Jonasson J, Forsum U. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS 2002; 110: 802–10
- Vasquez A, Jakobsson T, Ahrne S, Forsum U, Molin G. Vaginal lactobacillus flora of healthy Swedish women. J Clin Microbiol 2002; 40: 2746–9
- Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol 2002; 46: 535–48
- Sakamoto M, Umeda M, Benno Y. Molecular analysis of human oral microbiota. J Periodontal Res 2005; 40: 277–85
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Jr, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol 2006; 72: 2837–48
- de Lillo A, Ashley FP, Palmer RM, Munson MA, Kyriacou L, Weightman AJ, et al. Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol Immunol 2006; 21: 61–8
- Antonio MA, Rabe LK, Hillier SL. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J Infect Dis 2005; 192: 394–8
- Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001; 67: 2578–85