Abstract
Extracellular DNA (eDNA) is a macromolecule copiously found in various natural microenvironments, but its origin and significance still remain partly mysterious phenomena. Here, the multifaceted origins of eDNA in bacterial biofilms are explored. The release of eDNA can follow a suicidal programmed bacterial apoptosis or a fratricide-induced death, under the control of quorum sensing systems or triggered by specific stressors. eDNA can be released into the extracellular space or as a free macromolecule or enclosed within membrane vesicles or even through an explosion of bubbles. eDNA can also be derived from host tissue cells through bacterial cytolytic/proapoptotic toxins or stolen from neutrophil extracellular traps (NETs). eDNA can alternatively be produced by lysis-independent mechanisms. Sub-inhibitory doses of antibiotics, by killing a fraction of bacteria, result in stimulating the release of eDNA. Even phages appear to play a role in favoring eDNA release. Unveiling the origins of eDNA is critical to correctly address biofilm-associated infections.
Introduction
In recent years, extracellular DNA (eDNA) has increasingly emerged as one of the main extracellular polymeric substances (EPS) composing the architecture of bacterial biofilms. Other EPS taking part in bacterial biofilm matrices include various types of extracellular and cytoplasmic moonlighting (Jeffery Citation2014) proteins, neutral and electrostatically charged exopolysaccharides, teichoic acids and lipids. Differing from eDNA, these further EPS are generally species-specific and often even strain type-specific. Conversely, the presence of eDNA in biofilms appears highly conserved and widely distributed even across taxonomically distant bacterial and fungal microorganisms (Martins et al. Citation2010; Okshevsky & Meyer Citation2015). While many important functional roles of eDNA in biofilms have been documented and nowadays appear well established, the physiological mechanisms that are implicated in eDNA release and exert control over its production have only partly been unveiled and only in a restricted number of bacterial species (Sarkar Citation2020). Contrasting with the apparent eDNA ubiquity in biofilms, it appears as if, during phylogenesis, each single bacterial species reinvented alternative and/or complementary routes and strategies to repurpose, release or transfer DNA outside the cells.
eDNA is a very versatile and multifunctional molecule, which provides multiple benefits to microbial cells, enabling bacterial survival under highly stressing environmental conditions. eDNA favors horizontal gene transfer, enabling the interexchange of genes encoding antibiotic resistance and virulence traits between bacteria (Molin & Tolker-Nielsen Citation2003). It promotes bacterial aggregation (Mlynek et al. Citation2020) and enables adhesion to highly hydrophobic surfaces, which are most unfavorable to bacterial colonization (Das et al. Citation2011). When bacteria turn sessile following adhesion to a substratum or, otherwise, undergo flocculation while being suspended in physiologic fluids, eDNA acts as an element of the biofilm architecture, conferring structural stability through its many interactions with other EPS (Montanaro et al. Citation2011a). eDNA has been described as acidifying the local milieu, promoting antibiotic resistant phenotypes (Wilton et al. Citation2015), and chelating and neutralizing cationic bactericidal molecules including antibiotics (Chiang et al. Citation2013) and antimicrobial peptides (Batoni et al. Citation2016), actively contributing to biofilm tolerance. It triggers and conditions the host immune response (Hemmi et al. Citation2000; Islam et al. Citation2001; Fuxman Bass et al. Citation2010). Moreover, it facilitates self-organization of bacterial biofilms, guides biofilm spreading (Gloag et al. Citation2013), represents a nutrient source during starvation (Finkel & Kolter Citation2001), and exhibits extracellular electron transfer capabilities (Saunders et al. Citation2020).
Formation of eDNA is a necessary step for bacteria in order to benefit from all these functions. This article aims at reviewing the mechanisms of eDNA production adopted by different bacterial species, especially those commonly involved in nosocomial infections. It attempts to offer an overall view of the numerous and complex paths that have recently been unveiled.
The origin of eDNA in human infections: not just a single source
A more complete frame for the interpretation of the origin of DNA present in the extracellular space of biofilms of human pathogens has emerged only in recent years. In an early phase, eDNA was thought not to be actively released by bacteria, but rather to derive from bacterial cells facing a process of natural death. With time it became more evident though that, at least in species such as Streptococcus pneumoniae, induction of natural competence could actively trigger cell lysis and DNA release from a subfraction of the cell population (Steinmoen et al. Citation2002). Over the last two decades, the extensive research work conducted on many bacterial species has started to shed more light on the processes leading to the presence of eDNA in the intercellular space, revealing a multitude of alternative and often redundant mechanisms that vary with the pathogenic species. The scheme in summarizes some of the main mechanisms implicated in eDNA production, which will be discussed in detail in the following sections.
Figure 1. An overview of the numerous sources and mechanisms leading to eDNA accumulation in the extracellular space. Bacterial death and release of eDNA can be achieved by a variety of alternative, species- and strain-specific, mechanisms under the control of quorum sensing systems or triggered by specific stressors. Legend: MØs, monocytes/macrophages; PMNs, polymorphonuclear granulocytes; LTH, leukotoxic hypercitrullination; DNAJ, Heat Shock Protein DnaJ; PYO, pyocyanin; AHL, long-chain N-acyl-homoserine lactones; PQS, quorum-sensing molecule Pseudomonas quinolone signal; HQNO, 2-n-heptyl-4-hydroxyquinoline-N-oxide; C.bc1, cytochrome bc1 complex; ROS, radical oxygen species; OMV, outer membrane vesicle; OIMV, outer-inner membrane vesicle; EOMV, explosive lysis outer membrane vesicle.
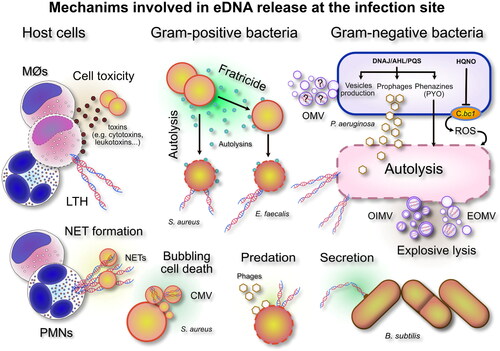
The most common systems adopted by bacteria to release DNA involve some type of programmed autolysis. In Gram-positive bacteria, this has been found often to occur through the production of peptides named autolysins. When being expressed, autolysins are capable of determining a sort of cell suicide or programmed apoptosis, by damaging the integrity of some vital cell components or, alternatively, to induce the death of nearby sister cells, causing a sort of fratricide (Montanaro et al. Citation2011b). Conversely, auto-intoxication and self-poisoning due to the production of toxic chemicals or a burst in the respiratory production of reactive oxygen species (ROS) has been described for Gram-negative bacteria such as Pseudomonas aeruginosa, which constitutes one of the best investigated bacterial models. The activation of prophages is another of the strategies pursued to achieve a programmed cell autolysis under the control of quorum sensing systems. The expression of phage endolysins can be associated with phenomena known as explosive lysis or bubbling cell death, in which case the autolytic disruption of bacterial cells generates membrane vesicles (MVs) with cytoplasmic content that may include also nucleic acids ().
Predation of bacteria by phages or other bacterial competitors is another mechanism leading to bacterial lysis and eDNA release. Moreover, sub-inhibitory doses of exogenous factors such as antibiotics have long been known to determine eDNA release. Interestingly, in a restricted number of species, alternative mechanisms have been observed where bacterial eDNA is secreted while cells are viable and maintain their integrity (Ibáñez de Aldecoa et al. Citation2017). Predation by other bacteria or bacteriophages represents a further circumstance in which bacterial DNA is released and can be used to enforce biofilm structure. Additionally, some antimicrobials and antibiotics capable of destabilizing the integrity of the bacterial wall have also been found to actively cause the release of eDNA and so favor biofilm production. Shields et al. (Citation2013) found that eDNA extracted from biofilms of different strains of staphylococci and streptococci tended to form a sharp band in agarose gel electrophoresis and exhibited a size >10 kb. However, the characteristics of bacterial DNA in the biofilm matrix likely reflect the diverse mechanisms used to produce eDNA and, while for certain bacteria, the sequence of eDNA has been found similar to that of genomic DNA (Qin et al. Citation2007; Rose et al. Citation2015), this has not always been the case for other bacteria (Jakubovics et al. Citation2013). For instance, in 2-day old biofilms of Helicobacter pylori, the comparison of DNA fingerprinting, performed by RAPD analysis on eDNA and intracellular DNA, showed substantial differences. Böckelmann et al. (Citation2006) reached similar conclusions analyzing the biofilm of a lotic bacterial strain, whose RAPD amplification profiles of the eDNA and genomic DNA revealed major similarities in the banding patterns, but also some significant differences.
In infected tissues eDNA is not uniquely of bacterial origin and restricted to the quantity of nucleic acids contributed by bacteria. Apart from programmed bacterial cytolysis, eDNA can also be produced from host cells such as leukocytes and other tissue cells. Indeed, host cells of the immune system such as leukocytes, but also other tissue cells have been found to represent an additional relevant source of extracellular nucleic acids released in the interstitial fluids. Following their activation, professional phagocytes are known to produce neutrophil extracellular traps (NETs), primarily consisting of extruded extracellular fibers of DNA (Zawrotniak & Rapala-Kozik Citation2013). Moreover, bacterial cytolytic toxins such as leukotoxins can determine the death of host cells and consequent release of high molecular weight DNA. As discussed in greater detail further on, while bacteria have been suggested to be potentially capable of harvesting and incorporating within their biofilm matrix the eDNA derived from these eukaryotic sources, some in vivo experimental evidence appears to exclude such a possibility. An important factor that should be kept in consideration is that some properties of bacterial eDNA, characterized by a high ratio of CG bases and a lower molecular size, differ from those of the eDNA contributed by host cells in terms of interactivity with host immune cells and their cell receptors.
From an evolutive point of view, bacteria therefore appear particularly skilled at developing complementary or alternative routes to take the greatest advantage of a fundamental organic molecule such as DNA. Either made available through a substantial sacrifice of individuals for the good of their community, or actively secreted bearing significant energetic costs, or recovered from the interstitial milieu where host cells have left it behind, eDNA can be recycled and repurposed from its original function (Mann et al. Citation2009; Thammavongsa et al. Citation2013; Ibáñez de Aldecoa et al. Citation2017). What is certain is that nothing is left to chance and fine mechanisms keep eDNA release under control, as well as its usage and degradation. The different routes of release and the molecular mechanisms that govern eDNA production are specifically addressed in the next paragraphs.
Bacterial cell lysis
An intriguing initial conundrum was1whether bacterial eDNA was in fact chromosomal DNA or, rather, consisted of newly synthesized oligonucleotides secreted on demand. The confirmation that, at least in the biofilm of some bacterial species, eDNA was similar/corresponded to genomic DNA, cleared the way for the hypothesis that DNA could derive from death of a subpopulation of bacterial cells. Based on current understanding, the lysis of bacterial cells is without doubts a major mechanism of eDNA release, broadly observed across both Gram-positive and Gram-negative pathogens. Bacterial lysis normally follows the expression of cytotoxic factors. This type of process is usually referred to as autolysis, when individual cells, through auto-poisoning, actuate a form of suicide (Rice et al. Citation2007) or an apoptosis-like programmed death. This ‘altruistic’ behavior of individual bacteria provides eDNA for the needs of the community. In contrast, cannibalism and allolysis take place when cytotoxic factors expressed by immune individuals are directed towards sibling cells (fratricide) or other species (predation) (Thomas et al. Citation2008, Citation2009). In Enterococcus faecalis isolates from orthopedic implant infections, the ability to produce autolytic enzymes proved crucial for expressing high levels of virulence (Arciola et al. Citation2008). The coordinated cell death in isogenic populations resulting in the cytolysis of a sub-population of genetically identical siblings for the benefit of the population appears a more general strategy not only assoiated with biofilm formation, but also with the initiation of competence and sporulation (Popp & Mascher Citation2019).
Among Gram-negative bacteria, P. aeruginosa is one of the most extensively investigated species. As illustrated in , in P. aeruginosa at least three main, apparently redundant, mechanisms have been identified that are potentially capable of lead ing to bacterial death and autolysis with consequent release of eDNA: (1) auto-poisoning by phenazines (Das & Manefield Citation2013; Sarkar Citation2020) and pyocyanin (PYO) (Das & Manefield Citation2012; Meirelles & Newman Citation2018); (2) auto-poisoning by 2-n-heptyl-4-hydroxyquinoline-N-oxide (HQNO) (Hazan et al. Citation2016); and activation of prophage endolysins (Turnbull et al. Citation2016; Li et al. Citation2019). In a recent review, Sarkar (Citation2020) explored in detail these diverse mechanisms. PYO is one of the most studied substances termed phenazines, which are produced by P. aeruginosa. Phenazines are redox-active dibenzo annulated pyrazines with two nitrogen atoms in the center ring having proton-coupled electron transfer properties. The toxicity of PYOs has been primarily associated with its redox activity as it can rapidly reduce molecular oxygen leading to the production of the reactive oxygen species superoxide, which is subsequently converted into H2O2 (Das & Manefield Citation2012). PYO mediated death is known to be induced under special environmental conditions, in particular nutrient depletion. Under other conditions such as reducing stress, PYO and phenazines seem to exhibit beneficial effects, serving as alternative electron acceptors and enabling cells to maintain redox homeostasis. The work of Meirelles & Newman (Citation2018) helped to elucidate the ‘double edged sword’ effects of PYOs. These authors observed that PYO production has predictable, distinct and striking cellular consequences and that these vary according to the physiological state of the producer. When cells are at a high cellular density, with high levels of carbon but electron acceptor-/oxidant-limited, PYO serves as an electron acceptor and avoids toxicity. Conversely, with starvation and low ATP levels, cells are not able to efficiently use all the mechanisms of defence from oxidative damage, which include an array of enzymes such as catalases, alkyl hydroperoxide reductases (Ahps), multiple thioredoxins and thioredoxins reductases (Ochsner et al. Citation2000) that are necessary to coup with PYO redox toxicity. This auto-poisoning mechanism should be interpreted as an altruistic sacrifice, where in front of the cost of individual cells, subsets of bacteria with PYO-tolerant characteristics thrive and take advantage of the eDNA released.
In recent years, apart from PYO, Hazan et al. (Citation2016) have produced evidence of another existing mechanism of auto-poisoning in P. aeruginosa, based on HQNO, a component of the quinolone signal system for quorum sensing. The authors demonstrated that, exhibiting intriguing similarities to programmed mitochondrial apoptosis in eukaryotic cells, HQNO causes cell death through the inhibition of the Qi site of the cytochrome bc1 complex (see ). Qi inhibition by HQNO disrupts the cytochrome c Q-cycle and leads to ROS accumulation to toxic levels and loss of membrane integrity, with consequent release of cytoplasmic content in the outer space (Hazan et al. Citation2016). Cytochrome c plays a fundamental role in the generation of ATP. Low concentrations of ATP are known to be associated with persister cells phenotypes. HQNO would thus potentially enhance tolerance to antibiotics both by causing the release of eDNA from autolytic cells (so strengthening the biofilm structure) and by promoting bacterial dormancy and persister cell formation (Hazan et al. Citation2016). It could be further speculated that lower ATP could also, in turn, create the preconditions that expose the bacterial cells to the damaging effects of PYO.
In clinical fluids such as cystic fibrosis sputum, HQNO can be produced in concentrations as high as 4 µM (Barr et al. Citation2015). However, the production of HQNO by P. aeruginosa varies depending on the environmental circumstances and, under cell culture conditions, up to ten times higher concentrations can be reached (Hazan et al. Citation2016). Despite the many P. aeruginosa protective mechanisms, these high levels of HQNO certainly represent a risk of autotoxicity. However, high HQNO concentrations are even more effective in killing and warding off other more sensitive Gram-positive bacteria inhabiting the same niches such as S. aureus. In addition to HQNO, a series of other exoproducts such as siderophores and rhamnolipids are typically produced by P. aeruginosa that are toxic to S. aureus. Therefore, in polymicrobial infections eDNA could also be generated from the lysis of the other coexisting bacteria. Up to 30% of the patients affected by cystic fibrosis have been found to be coinfected by these two dominant pathogens, which therefore appear to generate successful associations. A variety of host and environmental factors have been demonstrated to promote the co-existence of P. aeruginosa-S. aureus (Price et al. Citation2020). Along with auto-poisoning, another mechanism that has emerged to cause P. aeruginosa death and consequent eDNA release is explosive cell lysis. Turnbull et al. (Citation2016) have documented as the activation of a cryptic prophage endolysin, respectively the Lys endolysin, which is encoded by lys gene within the R- and F-pyocin genes cluster, can cause a form of violent cell death termed explosive cell lysis. The authors reported that, although R- and F-pyocins are known bacteriocins potentially implicated in cell lysis, only the pyocin endolysin Lys is required for eDNA production via explosive cell lysis. Lys has been hypothesized to translocate to the periplasm and degrade the peptidoglycan, thus enabling the release of the pyocins (Nakayama et al. Citation2000). During the lytic process, in addition to direct dispersion of cytoplasmic material in the outer space, Turnbull et al. (Citation2016) reported the biogenesis of MVs from the vesicularization of shattered membrane fragments. Toyofuku et al. (Citation2019) recently reviewed the origins and the content of MVs. Two main types of MVs were described as generated from explosive cell lysis, respectively outer-inner membrane vesicles (OIMV) and explosive lysis outer membrane vesicles (EOMV). Both types of MV would contain cytoplasmic material such as nucleic acids and proteins. In Gram-negative bacteria outer membrane vesicles (OMV) can be produced also by blebbing in the absence of cell death and not uniquely through cell lysis. However, with the exclusion of plasmid DNA, the presence of chromosomic DNA seems more likely associated uniquely to EOMV (Toyofuku et al. Citation2019). Thus, cell lysis-derived eDNA would include both DNA directly released in the outer space and DNA entrapped within MVs ().
In 2017, Ibáñez de Aldecoa et al. reviewed the literature on the mechanisms of eDNA release, which are observed also in other Gram-negative species that are not exclusively of clinical interest. In 8 out of the 15 species examined, eDNA was found to be released through lytic mechanisms of either lysis or autolysis (Acinetobacter calcoaceticus, Campylobacter jejuni, Caulobacter crescentus, Neisseria meningitides, Pseudomonas chlororaphis, Pseudomonas stuzeri and Sherwanella oneidensis in addition to P. aeruginosa). However, for up to a third of these species, the mechanism was reported to be still unknown.
A surprising lack of information concerns also a species as common and investigated as Escherichia coli, for which natural competence so far has not yet been described. Numerous toxin-antitoxin (TA) systems are known for E. coli, but little is known on their respective roles and their capacity to determine programmed cell death. The toxin HipA is member of the phosphatidylinositol 3/4-kinase superfamily and takes part with the antitoxin HipB to the HipBA TA system. Interestingly, Zhao et al. (Citation2013) reported an association between HipA and the enhancement of biofilm formation through DNA release. Nonetheless, in view of the variations among distinct strain types and the limited information on the other existing TA systems, more extensive studies are warranted to finally identify the mechanisms that are contributing to DNA release in this species.
As far as Gram-positive bacteria are concerned, Ibáñez de Aldecoa et al. (Citation2017) reported the documented involvement of lytic mechanisms of eDNA release in just 4 out of the 17 species considered, respectively Enterococcus faecalis, S. aureus, S. epidermidis () and S. pneumoniae. For seven of the species investigated there was still no available information on the mechanism involved in eDNA release and Streptococcus mutans had been described uniquely to release small size DNA fragments within MVs (Liao et al. Citation2014). The release had been observed to occur by growing cells during early biofilm formation, in the absence of lytic processes. However, shortly after, Jung et al. (Citation2017) reported on the role of the autolysin A (AtlA), confirming in a wild-type strain that eDNA release mediated by bacterial cell lysis is required for biofilm initiation and maturation. Thus, experimental evidence supports the co-existence of at least two alternative mechanisms for eDNA release in S. mutans.
In many Gram-positive pathogens, autolysis and allolysis have been found to be accomplished through the expression of well-identified bacteriocins/autolysins. Nonetheless, in many cases the molecular mechanisms are far from being totally understood. In E. faecalis, eDNA release and biofilm formation are achieved by a fratricidal mechanism, where death of sibling cells is actuated by isogenic cells within the same population by the primary N-acetyl glucosaminidase, autolysin AtlA (Thomas et al. Citation2009). Two extracellular peptidases were found to play an important antagonistic role in regulating the lytic activity of AtlA, respectively: (1) a gelatinase (GelE), which mediates cell death and eDNA release through proteolytical modification of AtlA, and (2) a serine protease (SprE), an immunity protein, inactive in the subpopulation of cells susceptible to lysis and capable of leading to an anti-lytic outcome.
Analogously, in Enterococcus faecium the autolysin AtlA, renamed AtlA(Efm), emerged among six putative autolysins as the major factor involved not only in bacterial lysis (and, thus, through the release of eDNA, in biofilm stabilization), but also in cell surface localization of the factor Acm. Acm mediates binding to collagen types I and IV (Paganelli et al. Citation2013).
In S. pneumoniae as in S. mutans, eDNA release is also ensured by mechanisms of allolysis/fratricide involving bacteriocins, where competent siblings target the lysis of non-competent cells to enable natural transformation (Steinmoen et al. Citation2002). However, the complexities of the pathway leading to competence differ, even between these two closely related species (Shanker & Federle Citation2017).
Autolysins have been proved to play an important role even in the case of some important clinically relevant staphylococcal species such as S. aureus and S. epidermidis. For S. aureus the main autolysin implicated in autolysis is AtlA, a bifunctional enzyme that undergoes proteolytic cleavage to yield two catalytically active proteins, an amidase (AM) and a glucosaminidase (GL). Both AM and GL functions are required for AtlA to be catalytically active and enable S. aureus to form a biofilm, demonstrating the significant contribution of AtlA-mediated lysis (Rice et al. Citation2007; Bose et al. Citation2012). Analogously, in S. epidermidis, the autolysin AtlE was found to have a role in the release of DNA and biofilm formation (Qin et al. Citation2007).
Bacterial eDNA from secretion
Over the years, cytolysis has progressively emerged not to be the only mechanism adopted by bacteria to release DNA in the outer space. Bacteria forming eDNA-containing biofilms, depending on the species and often on the strains, have occasionally been found to rely also on other alternative mechanisms of eDNA production, which are independent of cell death and lysis. One such mechanism is lysis-independent DNA secretion. Connected with natural cell transformation and competence functions, eDNA secretion is observed sometimes to co-exist with autolysis and allolysis in the same strain. Under such circumstances, eDNA secretion does not appear to cover a redundant function. Rather, it is implicated in the selective release of eDNA at different phases of the cell cycle and of biofilm maturation, being generally controlled by quorum sensing systems and influenced by external stimuli inducing the activation of competence.
This holds true for E. faecalis, an opportunistic pathogen that has previously been described to be capable of releasing eDNA through well-defined fratricide mechanisms. Nonetheless, lytic mechanisms are the source of eDNA for biofilm strengthening only at a late stage, i.e. in mature biofilms older than 24 h. This does not appear to occur during the early phases of biofilm formation, when visual examination under SEM and bulk biochemical assays would exclude the fact that significant lytic processes are taking place (Barnes et al. Citation2012). Conversely, during the initial stage, eDNA was found to be localized at the site of the septum of some cells as if a subpopulation of metabolically active cells could secrete/extrude eDNA in a way that is independent of cell lysis. The authors therefore reported the existence of a new non-lytic mechanism of DNA release. During the first 4 h of biofilm formation, they estimated a 3-Log increase in the ratio of eDNA per cell with respect to bacteria in the planktonic phase. The authors also reported that, during the first 4 h of biofilm growth, eDNA was primarily found in two distinct structures observable in the extracellular matrix, respectively in intercellular string-like structures that they defined yarns, and in the thick matrix surrounding the cells that were termed sweater (Barnes et al. Citation2012). Both yarns and sweater structures co-exist in individual biofilm samples. On the contrary, cell lysis would generate irregular masses of intracellular debris, these also including the nucleic acid component. Among the mechanisms capable of explaining this type of eDNA secretion occurring exclusively during biofilm formation, Barnes et al. (Citation2012) hypothesized the involvement of the conjugation apparatus or an as-yet-uncharacterized DNA export system.
In Neisseria gonorrhoeae, secretion of chromosomal DNA has been found to be associated to a type IV secretion system (TFSS) implicated in bacterial transformation. Salgado-Pabón et al. (Citation2010) observed that, within the cell population, gonococcal variants that produce type IV pili release larger quantities of DNA than non-piliated variants. Piliated strains would produce the largest quantity of DNA secretion in late log-phase growth and DNA release would not occur through autolysis. Moreover, secreted DNA would be in the form of single-strand DNA protected at the 5′ end from 5′–3′ exonuclease digestion (Salgado-Pabón et al. Citation2007). Secretion would occur between the 2-h and 2.5-h time points and precede the onset of the stationary/death phase during which bacteria would switch to cell lytic processes.
Nontypeable Haemophilus influenza (NTHI) has recently been the object of enlightening investigations on the machinery involved in the secretion of DNA. In NTHI, Jurcisek et al. (Citation2017) found that, in a subpopulation of NTHI cells, the transit to the outer space of DNA (and with it also of DNABII DNA-binding proteins, in particular the DNA-binding protein integration host factor, IHF) occurs through two steps. The first passage from the bacterial cytoplasm to the periplasm, across the inner membrane, takes place via the inner-membrane pore complex (TraCG), which is homologous to type IV secretion-like systems. Once across the inner membrane and having reached the periplasm, both DNA and DNABII proteins can exit to the outer space, being released into the environment through the ComE pore (the same ComE secretin through which the NTHI type IV pilus is expressed). Overall, ComE has therefore emerged as being involved in the formation of a type IV pilus, but also in the exportation and release of DNA and proteins in the outer space for biofilm formation and in the import of DNA for transformation (Krüger & Stingl Citation2011). Much remains to be elucidated on how these different functions are finally orchestrated (Seifert Citation2017).
Zafra et al. (Citation2012) found that in B. subtilis also the release of eDNA, corresponding to whole genomic double-stranded DNA, is linked to competence and occurs in the absence of cell lysis. For the bacterial strain investigated, named strain 3610, a peak of eDNA release was observed in the late exponential-phase, but this peak was not confirmed in other laboratory strains.
As anticipated earlier, in E. coli the mechanisms involved in eDNA release are mostly obscure. In 2010, Sanchez-Torres et al. investigated the functions of control of the global regulator H-NS on eDNA production and were led to postulate the existence of a release via direct eDNA secretion from living cells. Indeed, while the deletion of the gene encoding the global regulator H-NS resulted in the suppression of eDNA production, a slightly decreased cell lysis and the increased vesiculation observed could not be the explanation for the reduced eDNA.
Thus, in various Gram-positive and Gram-negative bacteria eDNA secretion appears to be linked to competence and to the DNA uptake machinery. The formation of complexes between single strand DNA (ssDNA) and the ComE secretin, its homologs found in other bacterial species (e.g. ComEA) or other unrelated proteins (e.g. single-stranded binding protein SsbB), has been hypothesized to act as a putative reservoir function. Under this hypothesis, periplasmic ComE/ComEA/SsbB-DNA clusters would be accumulated and stored until needed and become available on demand. This periplasmic accumulation could serve not only for transformation but also as a food source (Seitz & Blokesch Citation2014). Moreover, periplasmic accumulation would support the hypothesis that, in Gram-negative bacteria, vesiculation could generate DNA-containing OMVs.
However, a very recent discovery extends knowledge on the diversified mechanisms of eDNA release. Rugose small colony variant (RSCV) isolates of P. aeruginosa are known to be hyperactive in biofilm production and cause difficult to treat chronic infections (Pestrak et al. Citation2018). Deng et al. (Citation2020) reported that, in a hyperbiofilm-forming RSCV named PAO1DwspF, biofilm aggregates are formed by two bacterial subpopulations with contrasting physiological characteristics and a segregated spatial distribution. Interestingly, PAO1ΔwspF was found to possess the unique ability to extrude DNA fragments from living bacteria as part of bolstering the biofilm structure. This is in contrast with eDNA in the biofilm of the PAO1 isogenic parental strain, which consists of intact bacterial DNA as expected in a bacterial species earlier known to release eDNA through explosive cell lysis. Thus, considerable diversity characterizes the mechanisms of eDNA production, even among strain types of the same species.
Bacterial eDNA release through MVs
In an earlier section, MVs were described as associated with cell lysis events. However, this is not always the case. Gram-negative (Kulp & Kuehn Citation2010) and, more recently, Gram-positive bacteria (Brown et al. Citation2015) have both been found to be capable of producing MVs, i.e. nanosized bodies in the range 20-300 nm, which are delimited by cellular membranes, which form on the surface of the bacterial cells and are released free into the outer space. MVs have been found to be heterogenous, varying in composition and expressing different functionalities and properties, depending on their different pathways of biogenesis (Nagakubo et al. Citation2020) and on their growth phase transition (Tashiro et al. Citation2010). Based on their different cargos, diverse important functions have been described for MVs in (1) bacterial signaling and quorum sensing (Dauros-Singorenko et al. Citation2018; Toyofuku Citation2019); (2) immunomodulation of the host response by stimulating or quenching the immune system through induction of host cell apoptosis and delivery of toxins (Kaparakis-Liaskos & Ferrero Citation2015); (3) resistance to antimicrobial substances by antibiotic binding and sequestration, (4) by degrading enzymes or by contributing to horizontal gene transfer (Vitse & Devreese Citation2020); and finally (5) biofilm formation.
As far as biofilm production is concerned, MVs intervene at different levels. In the biofilm of some bacterial species, they represent a conspicuous fraction of the extracellular matrix and its EPS. About 20% of the whole matrix proteome of P. aeruginosa biofilms has been found to consist of MV-associated proteins (Couto et al. Citation2015). These proteins include among others: cytoplasmic molecules with moonlighting functions such as carbohydrate metabolic proteins and lipid metabolic proteins; proteins with defensive/protective functions such as SOS proteins and phage attack proteins; virulence factors such as host attack proteins and toxins (e.g. cytotoxins); and autolysins (Dineshkumar et al. Citation2020). Bacteriocins/autolysins secreted within MVs can eventually be used to induce the lysis in neighboring bacteria, leading to the release of nutrient molecules and eDNA. In addition to proteins, depending on the biogenesis pathway, the cargo of MVs has also been reported to include, among other molecules, lipids and nucleic acids. Nucleic acids in MV cargo comprise RNA (miRNA, mRNA and rRNA), but also single- and double-strand eDNA (Kadurugamuwa and Beveridge Citation1995), potentially involved in horizontal gene transfer as well as in biofilm formation. Furthermore, MVs produced by biofilm cells of Helicobacter pylori were suggested to prevent eDNA degradation by nucleases, provide a bridging function between eDNA strands on MV surfaces and mediate aggregation (Grande et al. Citation2015).
As shown in , there are distinct types of MVs. They can be produced through different biogenesis pathways, and this is reflected by a different cargo composition. Toyofuku et al. (Citation2019) reported as still open to debate the fact that OMVs, generated by blebbing and in absence of cell lysis events, may contain eDNA and, with current knowledge, it is difficult to clearly explain the transfer to the periplasm of genetic material leaching from the cytoplasm, without expecting the subsequent death of the bacterial cell. Conversely, when phage-derived endolysins degrade the proteoglycan cell wall and lead to explosive cell lysis, the so generated explosive lysis outer membrane vesicles (EOMVs) and, even more, the explosive lysis outer-inner membrane vesicles (EOIMVs) are likely to contain cytoplasmic material and, with it, eDNA. A similar circumstance occurs when Gram-positive species, for instance Staphylococcus spp. such as S. aureus, B. subtilis, S. pneumonia, Listeria monocytogenes and S. mutans, generate cytoplasmic membrane vesicles (CMVs) (Liao et al. Citation2014). In B. subtilis, the expression, under DNA stress conditions, of an endolysin encoded by a defective prophage has been found to weaken the structural integrity of the cell wall peptidoglycan (Toyofuku et al. Citation2017). CMVs were found to originate from the protrusion of the cell membrane through the holes caused by the endolysin to peptidoglycan. In Gram-positive cells, a thicker cell wall structure would avoid the swelling and explosive cell lysis characteristic of Gram-negative cells such as P. aeruginosa, where MVs were seen to result from the fusion of shattered membranes rather than from protrusion through the wall and pinching off. Through similar mechanisms, DNA-damaging agents and antibiotics inducing the SOS response such as ciprofloxacin were found to trigger CMV formation, but only in lysogenic S. aureus strains (Andreoni et al. Citation2019) with a prophage dependent-mechanism. Conversely, antibiotics such as β-lactam antibiotics (flucloxacillin and ceftaroline) exhibited also a different route of CMV production by directly weakening the peptidoglycan layer, thus evidencing alternative mechanisms of CMV production. The content of eDNA was found to be greater for CMVs when blebbing was caused by phage lysis.
After earlier investigations demonstrating the formation of outer-inner membrane vesicles (OIMVs) by the protrusion of both outer and plasma membranes in the antarctic bacterium Shewanella vesiculosa M7T, Pérez-Cruz et al. (Citation2015) provided some evidence that the production of OIMVs containing eDNA could even occur in strains of other clinically relevant species, including N. gonorrhoeae, P. aeruginosa and Acinetobacter baumannii, with a possible involvement in lateral gene transfer. Nonetheless, eDNA-containing OIMVs were found to represent only a minimal proportion of MVs, in the range 0.2-1.2%, depending on the strains. Furthermore, OIMVs have been well documented by ultrastructural studies conducted by Cryo-TEM. Nonetheless, the mechanisms that would trigger OIMV formation, causing a loss of integrity of the cell wall, and enable the protrusion of both inner and outer membranes with a portion of cytoplasmic content has not been still totally clarified. Even more obscure is the evolution of the fate of bacterial cells losing portions of their genomic DNA.
Overall, eDNA production through MV secretion has been well documented in both Gram-negative and Gram-positive bacteria. The presence of DNA has been detected in the cargo of different types of membrane vesicles, even if, in MVs generated consequent to cell lysis or through mechanisms involving prophagic endolysins, greater quantities of nucleic acid have been reported. MVs appear to be a critical junction between routes of different bacterial functions and much remains to be understood regarding the modalities controlling their cargo. Certainly, MVs provide a form of eDNA transfer that is protected from the degradation by nucleases (Grande et al. Citation2011). They not only take physical part in the structure of biofilms, contributing their cargo of eDNA and proteins, but also interact with free eDNA molecules in the extracellular space promoting aggregation. Much still remains to be uncovered on the half-life in the interstitial milieu and on the further evolution of MVs. Their fate can certainly be that of being incorporated and contributing to the biofilm, but it can also be that of fusion with the membranes of other adjacent prokaryotic cells in order to accomplish cell transformation, cell signaling or delivery of bacteriocins. A further important option is that of entering eukaryotic host cells and explicating pathogenic functions by hijacking host defenses.
Host cells and tissues eDNA
An important point when examining the different sources of eDNA in bacterial biofilms is that, while in biofilms cultured under artificial in vitro conditions, the only source of DNA is represented by bacteria, under real clinical conditions a proportion of EPS, DNA among them, is provided by host cells and tissues. In this regard, the role played by human extracellular matrix proteins such as fibronectin in S. aureus bacterial aggregations and biofilm formation is an example. Indeed, fibronectin can form a bridge and connect fibronectin-binding proteins, namely FnBPA and FnBPB, expressed on the outer surface of staphylococcal cells (Speziale & Pietrocola Citation2020). Adhesins such as FnBPs are virulence determinants and represent an adaptation of the pathogen to the physiological environment of the host, exploiting the wealth of molecules available in the interstitial milieu. Among other polymeric substances, DNA of host provenance is certainly a molecule of great potential value both as a source of nurture as well as a component that can be integrated into the biofilm to enforce its architecture and enhance its shielding functions. Indeed, different in vitro investigations have shown that supplementation with exogenous DNA can partially or totally restore some lost functionalities in bacterial biofilms treated with DNase (Carrolo et al. Citation2010; Harmsen et al. Citation2010; Das et al. Citation2011; Chiang et al. Citation2013; Wongkaewkhiaw et al. Citation2020). Nonetheless, there is some evidence that, under real in vivo conditions, in infections caused by P. aeuruginosa, the majority of eDNA is found external to the biofilm, keeping it confined in the peripheral space, and derived from the host rather than from bacteria (Alhede et al. Citation2020).
Except under particular pathological conditions such as malignancies and infections, the presence of eDNA in physiologic interstitial fluids is generally low. In bacterial infections, the release of DNA from host cells can be associated to different phenomena. These include: (1) the death of host tissue cells/leukocytes for the action of bacterial cytolytic or proapoptotic toxins; (2) tissue damage derived from an excess of inflammatory reaction; (3) production of neutrophil extracellular traps (NETs) by host leukocytes.
Pathogens such as S. aureus, a main causative agent of nosocomial infections, are known to express several toxins and pore forming cytolysins that are capable of causing the death of host tissue cells and of effector cells of the immune system (Arciola et al. Citation2018; Campoccia et al. Citation2019). The S. aureus virulon comprises numerous toxins, among them PSMα3, δ-Toxin and other leukocidins, bi-component β-barrel pore-forming toxins including γ-hemolysin and Panton Valentine leukocidin (PVL) (Vandenesch et al. Citation2012; Arciola et al. Citation2018; Tromp & van Strijp Citation2020). In addition to the secretion of cytolytic toxins, S. aureus can degrade eDNA by the combined action of two enzymes, nuclease and adenosine synthase A, producing the powerful pro-apoptotic metabolite dAdo, capable of affecting the viability of host leukocytes (Thammavongsa et al. Citation2013; Papayannopoulos Citation2018; Campoccia et al. Citation2019). Moreover, S. aureus has been found to possess further mechanisms leading to cells apoptosis and autophagocytosis. For instance, this species can efficiently internalize into host cells such as bone cells and, thereby, upregulate the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which, in turn, induces osteoblast apoptosis with consequent bone-tissue damage, through the activation of caspase 8 (Alexander et al. Citation2003). The special multifaceted ability of S. aureus to modulate the expression of virulence factors to prevent elimination through programmed cell death pathways and evoking apoptosis, necroptosis, and pyroptosis has recently been reviewed by Soe et al. (Citation2021).
Analogous cytotoxins have been identified in other pathogens. For instance, in P. aeruginosa, rhamnolipid has been reported to determine the lysis of neutrophil cells by acting as a biosurfactant ( Van Gennip et al. Citation2009). The ExoU cytotoxin and exolysin (ExlA) have also been described as two important exotoxins of P. aeruginosa (Tamura et al., Citation2004; Basso et al. Citation2017). Other examples of toxins expressed by other pathogens and capable of causing host cell death include the α-hemolysin of E. coli, the cytolysin of Vibrio cholerae, and the listeriolysin O of Listeria monocytogenes (Steinmoen et al. Citation2002). The action of cytolytic, necrotizing or pro-apoptotic mechanisms/factors can result in cell death and contribute to the release of substantial quantities of nucleic acids.
However, in the presence of an excess of inflammatory response, tissue disruption and cellular damage can also derive from the disproportionate activation of effector cells of the immune system. This can be triggered either by the presence of foreign bodies such as implant materials and their wear debris or by unresolved biofilm-associated chronic infections. Both of these conditions can determine frustrated leukocyte phagocytosis and release into the outer space of potentially harmful chemicals, normally confined within phagosomes (Arciola et al. Citation2018). Thus, the large arsenal of molecules usually exploited to contain invading microorganisms or generally directed against foreign bodies poses a risk of collateral damage to surrounding tissues (Kzhyshkowska et al. Citation2015). Under normal conditions, the successful resolution of acute inflammation is characterized by the apoptosis of neutrophil cells and their engulfment by macrophages. Conversely, in chronic bacterial infections, neutrophil cells are thought to undergo necrosis rather than apoptosis (Cox et al. Citation1995).
A third important mechanism of eDNA extrusion consists of the release by neutrophil cells of NETs, i.e. extracellular meshes composed of eDNA in the form of nuclear chromatin and granule proteins. NETs granule proteins include enzymatically active proteases such as neutrophil elastase, myeloperoxidase (MPO) and cathepsin G, and other antimicrobial peptides (e.g. histone H3) that bind, trap and kill bacteria. The NETome has been described as consisting of up to 24 different proteins (Urban et al. Citation2009). First described by (Brinkmann et al. Citation2004), the production of NETs was only later recognized as a specific and regulated form of cell death also known as NETosis (Fuchs et al. Citation2007) and reported to depend on the generation of reactive oxygen species (ROS) by NADPH oxidase (Steinberg & Grinstein Citation2007). NET formation was shown to be triggered by the presence of microorganisms as well as by biochemical stimuli such as phorbol myristate acetate (PMA) (Takei et al. Citation1996) or cytokines such as interleukin-8 (IL-8). Rapid killing of neutrophils by PMA accompanied by changes different from typical apoptosis and necrosis was reported by Takey et al. as early as1996. Despites the many efforts made to finely characterize all the triggering stimuli, the molecular mechanisms respectively implicated in NET formation and the sources of the DNA released (nuclear, mitochondrial or both) in order to reconduct the entire phenomenon to a single well-defined process, there are still many disputed points that remain unresolved (Boeltz et al. Citation2019). The nature of the mechanisms of eDNA extrusion leading to NET formation and the definition of NETosis have repeatedly been reported as mistakenly confused. In an attempt of clarification, Konig and Andrade (Citation2016) critically reviewed all the known mechanisms and processes of eDNA extrusion by neutrophils. In their reappraisal, they distinguished a properly defined NETosis from other similar processes involving eDNA release, based on the triggering stimuli, the activation of distinct biochemical pathways, the presence of hypercitrullination, and the antimicrobial effector function. Proper NETosis was defined as a form of programmed cell death intended to contrast the presence of pathogens and NETs as part of a defence mechanism aimed at immobilizing and neutralizing pathogenic microorganisms such as bacteria, fungi, and viruses. For Konig and Andrade (Citation2016), differing from other NETs mimicking conditions, NETosis was NADPH oxidase-dependent and was not associated with hypercitrullination. Conversely, DNA extrusion could also be triggered by bacteria, through the production of cytolysins. The authors defined this form of eDNA release as leukotoxic hypercitrullination (LTH). Triggered by pore-forming pathways and equivalent signals, LTH would involve calcium-dependent hyperactivation of citrullinating enzymes such as peptidylarginine deiminase type 4 (PAD4), protein hypercitrullination, and neutrophil death. Peptidylarginine deiminase activity would be involved in chromatin decondensation. LTH would not consist of a host defensive strategy of the immune system, but rather in a bacterial strategy to elude the host immune response.
In contrast, Yousefi et al. (Citation2019) reviewed the concept of NETosis and NET formation and argued that NET formation and neutrophil necrotic death with DNA release should be considered as independent phenomena. They discussed the evidence supporting NET formation in viable neutrophils and the molecular mechanisms and intracellular events leading to NET formation through mitochondrial DNA release. This type of NET formation, also observed in other types of granulocytes as well as in lymphocytes, would not involve cell death. It would occur rapidly, i.e. in a matter of minutes, and should be treated differently from the events of programmed necrosis, occurring with a 2-h delay. This view is aligned with the previous explicit recommendations of the Nomenclature Committee on Cell Death (NCCD) (Galluzzi et al. Citation2018) to avoid the use of the term NETosis, when no experimental evidence in support of cell death is available. Additionally, NCCD called for additional studies to address the still unclear contribution of necroptosis to NET extrusion and NETotic cell death. Based on the available evidence, NETotic cell death was defined as a ROS-dependent modality of regulated cell death, which is restricted to cells of hematopoietic derivation and associated with NET extrusion. Indeed, initially thought to be confined to neutrophils, the production of DNA extracellular traps (ETs), was found not to be exclusive of neutrophils, but to extend to other leukocytes, eosinophils (EETs) and to monocytes/macrophages (MoETs/METs) (Guimarães-Costa et al. Citation2012; Gómez et al. Citation2021). Analogously to the judgment expressed for NETosis, NCCD discouraged the use of the term ETosis (Galluzzi et al. Citation2018).
Apart from the many, still open, debates on the nomenclature and on the different characterization of the mechanisms, NET formation may be considered a complex and only partly understood phenomenon. Its underlying mechanisms vary as a function of the triggering stimuli that include a broad range of factors. As suggested by Castanheira & Kubes (Citation2019), it could be more appropriate simply to cautiously distinguish between lytic and non-lytic NET formation. In non-lytic NET formation, mitochondrial DNA most likely represents the source of the eDNA, whereas nuclear DNA release would involve or be the consequence of cell death. TLRs, complement receptors and lipopolysaccharide-activated platelets would be implicated in non-lytic NET formation directed to counter the presence of bacteria. Conversely, bacterial pore-forming leukocidins such as PVL and γ-hemolysin that are expressed by S. aureus would be capable of causing both NET formation and cell death, although it remains unclear if, in this context, NETs are simply a consequence of neutrophil lysis. Intriguingly, the induction of NETs appears to represent an advantage for these bacteria (Bhattacharya et al. Citation2018) and favor their persistence in the tissues rather than representing a real threat. Differing from planktonic bacteria, S. aureus biofilms would rapidly skew neutrophils toward NET formation through the combined activity of PVL and γ-hemolysin. By eliciting this response, S. aureus would persist in the host tissues, as the result of the antimicrobial activity of NETs would be ineffective at clearing biofilm bacteria. Very recently Mazzoleni et al. (Citation2021) described for PVL the existence of an alternative NETosis process targeting mitochondria. PVL would colocalize with mitochondria, enhance the production of reactive oxygen species from these organelles and cause the ejection of chromatin fibers independently of the NADPH oxidase oxidative burst. Conversely, the pore-forming cytolytic leukotoxin GH (LukGH), differing from PVL, was found neither to prime human neutrophils for increased ROS production nor to enhance the uptake of S. aureus. Nonetheless, LukGH was found to be capable of promoting the release of NETs, even if they merely ensnared but did not kill the bacteria (Malachowa et al. Citation2013).
Hypothetical advantages for bacteria inducing NET formation could derive from the contribution of NET DNA to the biofilm architecture as well as from the conversion of this DNA to the pro-apoptotic metabolite dAdo (Thammavongsa et al. Citation2013; Arciola et al. Citation2018; Papayannopoulos Citation2018; Missiakas & Winstel Citation2021). However, in in vivo chronic infections caused by P. aeruginosa such as those observed in lung tissues of cystic fibrosis patients, Alhede et al. (Citation2020) reported that bacterial biofilms seem incapable of triggering the release of NETs by PMNs. Moreover, eDNA released following PMNs necrosis would not apparently integrate and become part of the in vivo biofilm architecture. Rather, it would constitute a sort of secondary matrix, externally lining the biofilm and possibly acting as a passive physical shield against antibiotics and phagocytes. Thanabalasuriar et al. (Citation2019) reported that in ocular biofilms by the same pathogen, high expression of the bacterial type-3 secretion system (T3S) results in NETosis, with NETs forming a barrier ‘dead-zone’, capable of confining the infection, between host cells and P. aeruginosa biofilms.
In view of the above observations, it would appear as if, in lung tissues of cystic fibrosis patients, the release of host eDNA was determined by the virulence mechanisms actuated by the bacteria rather than representing, at least under these circumstances, a functional and effective host defence mechanism. The release of host DNA would increment the external protection to an organized biofilm underlayer. However, in cystic fibrosis as well as in ocular infections, the sacrifice of the leukocytes would generate a line of confinement capable of slowing down the spreading of bacteria. Moreover, bacterial DNA has some peculiarities compared with human DNA, being rich in hypomethylated ‘CpG’ repeats (Itagaki et al. Citation2011). Intracellular leukocyte receptors such as TLR9 have long be known to recognize unmethylated CpG motif characteristic of bacterial DNA (Hemmi et al. Citation2000; Otto Citation2008). Thus, the externally lining layer of host DNA could possibly mask this interaction, competing with bacterial eDNA in the uptake into endosomal compartments.
Overall, the role of NETs and of the DNA released by host cells remain difficult to interpret as far as their contribution to bacterial biofilms is concerned. Many alternative physiological mechanisms are emerging as implicated in the release of NETs. They do pinpoint a function of NETs in the host defence against microorganisms and parasites. However, some of them are voluntarily actuated by some pathogens, which appear to control and even trigger eDNA release rather than suffer its action. Abundant evidence highlights the fact that specialized opportunistic pathogens such as, for instance, S. aureus have evolved multiple mechanisms to avoid harm by the many antimicrobial substances associated with NETs (neutrophil elastase, MPO, cathepsin G, and histones such as H3). They do induce the release of nucleic acids and, once their bactericidal potential is neutralized, their reward would be expected to consist in survival from the neutrophil trap and even in the DNA itself derived from the inactivated traps. However, the in vivo observations in a murine model and in human lung tissues of cystic fibrosis patients consistently indicate that neutrophil-derived DNA does not co-localise with P. aeruginosa biofilms (Alhede et al. Citation2020) as contrastingly do neutrophil elastase and citrullinated H3.
Under normal conditions (i.e. in the absence of a foreign body, in subjects that are not affected by genetic diseases such as cystic fibrosis and in non-immunocompromised patients), the innate and cell mediated immune response of the host is generally effective in warding off the majority of the infections and preventing the establishment of recalcitrant biofilm infections. However, when an imbalance of the immune defenses favors invading opportunistic pathogens, these can skew the antibacterial action for which NETs are intended and turn a risk in an opportunity. Ríos-López et al. (Citation2021) have recently reviewed the multiplicity of mechanisms that pathogens have developed to escape the bactericidal action for which NETs are intended.
Biomaterial-triggered NETs
An important aspect concerning implant associated infections is that neutrophils actively interact with biomaterial surfaces (Selders et al. Citation2017). Neutrophils are rapidly recruited at the site of implantation and wherever biomaterial surfaces come in direct contact with blood. Since the early stages, these leukocytes play a fundamental role in orchestrating the tissue response to foreign body surfaces and as such to biomaterial surfaces. An only recently emerged activity of these effector cells is their ability of priming/preconditioning biomaterial surfaces. It has long been known that matrix proteins present in the physiological fluids, including components of the humoral immune response (e.g. factors of the coagulation and complement cascade), nearly instantaneously adsorb on biomaterial surfaces generating a proteinaceous conditioning film. Immediately after this interaction with proteins also known under the name of Vroman effect, cell-mediated host immune defenses react to the biomaterial surface of the implant, recognizing it as a foreign body. Depending on physical and chemical cues, the biomaterial surface can elicit leukocyte activation and NET extrusion, the latter process resulting in an additional conditioning of the outer surface, which eventually becomes coated with eDNA. All this has important implications for the pathogenesis of implant infections. First leukocyte activation following the interaction with implant materials can result in metabolic exhaustion and in a reduced capacity of neutrophils to release ROS and kill bacteria, resulting in a niche of local immune depression at the interface of implant with periprosthetic tissues. Local immune depression is one of the possible reasons enabling typically saprophytic bacterial species to emerge as opportunistic pathogens in the presence of implant materials (von Eiff et al. Citation2006). Second, it is likely that the presence of NETs eDNA on the biomaterial surface can electrostatically attract and entrap bacteria, facilitating microbial adhesion and colonization. In this perspective, even if it is still unproven that eukaryotic eDNA can always be effectively incorporated in the extracellular matrix of bacterial biofilms, NETs could certainly represent nuclei of bacterial aggregation and, in the absence of a solid substrate, of flocculation.
eDNA in orthopedic infections
Orthopedic infections deserve special attention, since more than a million total hip and knee arthroprostheses are implanted each year in US, and this number is expected to increase as population ages (Singh et al. Citation2019). In periprosthetic infections, bacterial contamination can occur in two ways: (1) through accidental contamination of the surgical incision by bacteria either from the patient's skin or mucosae, the healthcare personnel, or the operating room environment, during surgery; (2) through hematogenous spread from a distant focus of infection, during the postoperative period.
The role of eDNA in periprosthetic orthopedic infections is of great interest. In a study aimed at investigating the relation between PIA and eDNA in a large collection of S. epidermidis clinical isolates from orthopedic infections, the presence in the biofilm matrix of the exopolysaccharide was found to correlate with that of eDNA. Isolates that did not produce PIA exhibited minimal levels of eDNA, while strains with significant PIA production showed high amounts of eDNA (Campoccia et al. Citation2011). Furthermore, eDNA was found to be a constant component of the various biofilm matrices produced by S. lugdunensis isolates from orthopedic implant infections (Ravaioli et al. Citation2020).
In orthopedic prosthetic joint infections by S. epidermidis, Zatorska et al. (Citation2018) showed a correlation between a high amount of eDNA in biofilms of bacterial clinical isolates and adverse clinical outcome. Therefore, quantification of bacterial eDNA may represent a predictive marker for the management of joint infections (Zatorska et al. Citation2018).
The excerpts mentioned foreshadow the potential clinical utility of an in-depth study of the origins of eDNA. In this connection, it must be emphasized that the field of orthopedic implants is one of the most burdened by biofilm-associated infections.
Conclusions
Although bacterial mechanisms of eDNA production are progressively being unveiled, they still warrant extensive investigative work. Directly originated by the action of noxious agents affecting bacterial integrity, induced by the activation of dedicated quorum sensing systems, or contributed by host cells, eDNA production has a critical impact on biofilm formation and the possibility of pathogen persistence in the environment as well as in the human body. For this reason, this ubiquitous molecule is increasingly being considered an ideal target for broad-spectrum anti-biofilm strategies aiming at disrupting biofilm integrity. However, eDNA susceptibility to enzymic degradation is generally limited to the very early stages of biofilm formation, when the polyanionic polymer has not yet undergone extensive complexation with other EPS, in particular amyloid and polycationic proteins, and exopolysaccharides. Thus, most advanced strategies are being directed to interfering with, preventing or disrupting the interactions of eDNA with other EPS, thus disabling or dismantling fully protected mature biofilms.
A better understanding of the origin of eDNA and on how controlled bacterial death can represent a real advantage for the bacterial population is of paramount importance. Alternative measures for prevention and treatment of biofilm-based infections that do not strictly target DNA have to be meticulously thought and assessed, carefully considering the risk of causing the release of eDNA and, thus, enhancing biofilm formation. For instance, this is a case of the much-discussed phage therapy. Therapies based on the use of phage viruses certainly represent a promising tool to partly obviate the problem of the current shortage of efficacious antibiotic treatments. However, phages are not only implicated in horizontal gene transfer through genetic transduction, but they can also establish symbiotic relationships that induce and strengthen biofilms (Pires et al. Citation2021). Furthermore, prophagic autolysins have themselves emerged among the mechanisms used by bacteria to release eDNA and thrive in hostile environments. Thus, an initial bactericidal effect obtained by experimental phage therapies should not be confused with a successful cure, and final complete and long-lasting remission of an infection should be proved after a prolonged follow-up, particularly in the presence of prosthetic devices.
Similar considerations can be made regarding the use of delivery systems based on the release of antibiotic or bactericidal substances intended to prevent biofilm formation on prosthetic surfaces. If the bactericidal action is not perfectly accomplished and a protracted subinhibitory release of antibiotic is perpetuated at the site of implantation, there is a real risk of promoting the formation of eDNA, consequently enhancing biofilm production instead of achieving a successful eradication of the infection.
In addition, there is a real need to gain a better comprehension of the role of eDNA derived from host cells in the perpetuation of chronic infections, especially those developing in the absence of a foreign body.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alexander EH, Rivera FA, Marriott I, Anguita J, Bost KL, Hudson MC. 2003. Staphylococcus aureus - induced tumor necrosis factor - related apoptosis - inducing ligand expression mediates apoptosis and caspase-8 activation in infected osteoblasts. BMC Microbiol. 3:5. doi:https://doi.org/10.1186/1471-2180-3-5
- Alhede M, Alhede M, Qvortrup K, Kragh KN, Jensen PØ, Stewart PS, Bjarnsholt T. 2020. The origin of extracellular DNA in bacterial biofilm infections in vivo. Pathog Dis. 78:ftaa018. doi:https://doi.org/10.1093/femspd/ftaa018
- Andreoni F, Toyofuku M, Menzi C, Kalawong R, Mairpady Shambat S, François P, Zinkernagel AS, Eberl L. 2019. Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob Agents Chemother. 63:e01439–18. doi:https://doi.org/10.1128/AAC.01439-18
- Arciola CR, Baldassarri L, Campoccia D, Creti R, Pirini V, Huebner J, Montanaro L. 2008. Strong biofilm production, antibiotic multi-resistance and high gelE expression in epidemic clones of Enterococcus faecalis from orthopaedic implant infections. Biomaterials. 29:580–586. doi:https://doi.org/10.1016/j.biomaterials.2007.10.008
- Arciola CR, Campoccia D, Montanaro L. 2018. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 16:397–409. doi:https://doi.org/10.1038/s41579-018-0019-y
- Barnes AM, Ballering KS, Leibman RS, Wells CL, Dunny GM. 2012. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio. 3:e00193–e00112. doi:https://doi.org/10.1128/mBio.00193-12
- Barr HL, Halliday N, Cámara M, Barrett DA, Williams P, Forrester DL, Simms R, Smyth AR, Honeybourne D, Whitehouse JL, et al. 2015. Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur Respir J. 46:1046–1054. doi:https://doi.org/10.1183/09031936.00225214
- Basso P, Ragno M, Elsen S, Reboud E, Golovkine G, Bouillot S, Huber P, Lory S, Faudry E, Attrée I. 2017. Pseudomonas aeruginosa Pore-Forming Exolysin and Type IV Pili Cooperate To Induce Host Cell Lysis. mBio. 8:e02250–16. doi:https://doi.org/10.1128/mBio.02250-16
- Batoni G, Maisetta G, Esin S. 2016. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta. 1858:1044–1060. doi:https://doi.org/10.1016/j.bbamem.2015.10.013
- Bhattacharya M, Berends ETM, Chan R, Schwab E, Roy S, Sen CK, Torres VJ, Wozniak DJ. 2018. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc Natl Acad Sci U S A. 115:7416–7421. doi:https://doi.org/10.1073/pnas.1721949115
- Böckelmann U, Janke A, Kuhn R, Neu TR, Wecke J, Lawrence JR, Szewzyk U. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol Lett. 262:31–38. doi:https://doi.org/10.1111/j.1574-6968.2006.00361.x
- Boeltz S, Amini P, Anders HJ, Andrade F, Bilyy R, Chatfield S, Cichon I, Clancy DM, Desai J, Dumych T, et al. 2019. To NET or not to NET: current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death Differ. 26:395–408. doi:https://doi.org/10.1038/s41418-018-0261-x
- Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One. 7:e42244. doi:https://doi.org/10.1371/journal.pone.0042244
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. doi:https://doi.org/10.1126/science.1092385
- Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 13:620–630. doi:https://doi.org/10.1038/nrmicro3480
- Campoccia D, Mirzaei R, Montanaro L, Arciola CR. 2019. Hijacking of immune defences by biofilms: a multifront strategy. Biofouling. 35:1055–1074. doi:https://doi.org/10.1080/08927014.2019.1689964
- Campoccia D, Montanaro L, Ravaioli S, Pirini V, Cangini I, Arciola CR. 2011. Exopolysaccharide production by Staphylococcus epidermidis and its relationship with biofilm extracellular DNA. Int J Artif Organs. 34:832–839. doi:https://doi.org/10.5301/ijao.5000048
- Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. 2010. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One. 5:e15678. doi:https://doi.org/10.1371/journal.pone.0015678
- Castanheira FVS, Kubes P. 2019. Neutrophils and NETs in modulating acute and chronic inflammation. Blood. 133:2178–2185. doi:https://doi.org/10.1182/blood-2018-11-844530
- Chiang WC, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 57:2352–2361. doi:https://doi.org/10.1128/AAC.00001-13
- Couto N, Schooling SR, Dutcher JR, Barber J. 2015. Proteome profiles of outer membrane vesicles and extracellular matrix of Pseudomonas aeruginosa biofilms. J Proteome Res. 14:4207–4222. doi:https://doi.org/10.1021/acs.jproteome.5b00312
- Cox G, Crossley J, Xing Z. 1995. Macrophage engulfment of apoptotic neutrophils contributes to the resolution of acute pulmonary inflammation in vivo. Am J Respir Cell Mol Biol. 12:232–237. doi:https://doi.org/10.1165/ajrcmb.12.2.7865221
- Das T, Krom BP, van der Mei HC, Busscher HJ, Sharma PK. 2011. DNA-mediated bacterial aggregation is dictated by acid-base interactions. Soft Matter. 7:2927–2935. doi:https://doi.org/10.1039/c0sm01142h
- Das T, Manefield M. 2012. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One. 7:e46718. doi:https://doi.org/10.1371/journal.pone.0046718
- Das T, Manefield M. 2013. Phenazine production enhances extracellular DNA release via hydrogen peroxide generation in Pseudomonas aeruginosa. Commun Integr Biol. 6:e23570. doi:https://doi.org/10.4161/cib.23570
- Das T, Sharma PK, Krom BP, van der Mei HC, Busscher HJ. 2011. Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: influence of ionic strength and substratum hydrophobicity. Langmuir. 27:10113–10118. doi:https://doi.org/10.1021/la202013m
- Dauros-Singorenko P, Blenkiron C, Phillips A, Swift S. 2018. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol Lett. 365: 1–9. doi:https://doi.org/10.1093/femsle/fny023
- Deng B, Ghatak S, Sarkar S, Singh K, Das Ghatak P, Mathew-Steiner SS, Roy S, Khanna S, Wozniak DJ, McComb DW, et al. 2020. Novel bacterial diversity and fragmented eDNA identified in hyperbiofilm-forming Pseudomonas aeruginosa rugose small colony variant. iScience. 23:100827. doi:https://doi.org/10.1016/j.isci.2020.100827
- Dineshkumar K, Aparna V, Wu L, Wan J, Abdelaziz MH, Su Z, Wang S, Xu H. 2020. Bacterial bug-out bags: outer membrane vesicles and their proteins and functions. J Microbiol. 58:531–542. doi:https://doi.org/10.1007/s12275-020-0026-3
- Finkel SE, Kolter R. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol. 183:6288–6293. doi:https://doi.org/10.1128/JB.183.21.6288-6293.2001
- Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. 2007. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 176:231–241. doi:https://doi.org/10.1083/jcb.200606027
- Fuxman Bass JI, Russo DM, Gabelloni ML, Geffner JR, Giordano M, Catalano M, Zorreguieta A, Trevani AS. 2010. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. JI. 184:6386–6395. doi:https://doi.org/10.4049/jimmunol.0901640
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. 2018. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25:486–541. doi:https://doi.org/10.1038/s41418-017-0012-4
- Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, et al. 2013. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 110:11541–11546. doi:https://doi.org/10.1073/pnas.1218898110
- Gómez RM, López Ortiz AO, Schattner M. 2021. Platelets and extracellular traps in infections. Platelets. 32:305–313. doi:https://doi.org/10.1080/09537104.2020.1718631
- Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. 2011. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol. 110:490–498. doi:https://doi.org/10.1111/j.1365-2672.2010.04911.x
- Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, Paolino D, Codagnone M, Muraro R, Stoodley P, et al. 2015. Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front Microbiol. 6:1369. doi:https://doi.org/10.3389/fmicb.2015.01369
- Guimarães-Costa AB, Nascimento MT, Wardini AB, Pinto-da-Silva LH, Saraiva EM. 2012. ETosis: a microbicidal mechanism beyond cell death. J Parasitol Res. 2012:929743. doi:https://doi.org/10.1155/2012/929743
- Harmsen M, Lappann M, Knøchel S, Molin S. 2010. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol. 76:2271–2279. doi:https://doi.org/10.1128/AEM.02361-09
- Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, Rahme LG. 2016. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr Biol. 26:195–206. doi:https://doi.org/10.1016/j.cub.2015.11.056
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. 2000. A toll-like receptor recognizes bacterial DNA. Nature. 408:740–745. doi:https://doi.org/10.1038/35047123
- Ibáñez de Aldecoa AL, Zafra O, González-Pastor JE. 2017. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front Microbiol. 8:1390. doi:https://doi.org/10.3389/fmicb.2017.01390
- Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. 2001. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 7:180–185. doi:https://doi.org/10.1038/84627
- Itagaki K, Adibnia Y, Sun S, Zhao C, Sursal T, Chen Y, Junger W, Hauser CJ. 2011. Bacterial DNA induces pulmonary damage via TLR-9 through cross-talk with neutrophils. Shock. 36:548–552. doi:https://doi.org/10.1097/SHK.0b013e3182369fb2
- Jakubovics NS, Shields RC, Rajarajan N, Burgess JG. 2013. Life after death: the critical role of extracellular DNA in microbial biofilms. Lett Appl Microbiol. 57:467–475. doi:https://doi.org/10.1111/lam.12134
- Jeffery CJ. 2014. An introduction to protein moonlighting. Biochem Soc Trans. 42:1679–1683. doi:https://doi.org/10.1042/BST20140226
- Jung CJ, Hsu RB, Shun CT, Hsu CC, Chia JS. 2017. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun. 85:e00252–17. doi:https://doi.org/10.1128/IAI.00252-17
- Jurcisek JA, Brockman KL, Novotny LA, Goodman SD, Bakaletz LO. 2017. Nontypeable Haemophilus influenzae releases DNA and DNABII proteins via a T4SS-like complex and ComE of the type IV pilus machinery. Proc Natl Acad Sci U S A. 114:E6632–E6641. doi:https://doi.org/10.1073/pnas.1705508114
- Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 177:3998–4008. doi:https://doi.org/10.1128/jb.177.14.3998-4008.1995
- Kaparakis-Liaskos M, Ferrero RL. 2015. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 15:375–387. doi:https://doi.org/10.1038/nri3837
- Konig MF, Andrade F. 2016. A critical reappraisal of neutrophil extracellular traps and NETosis mimics based on differential requirements for protein citrullination. Front Immunol. 7:461. doi:https://doi.org/10.3389/fimmu.2016.00461
- Krüger NJ, Stingl K. 2011. Two steps away from novelty-principles of bacterial DNA uptake. Mol Microbiol. 80:860–867. doi:https://doi.org/10.1111/j.1365-2958.2011.07647.x
- Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 64:163–184. doi:https://doi.org/10.1146/annurev.micro.091208.073413
- Kzhyshkowska J, Gudima A, Riabov V, Dollinger C, Lavalle P, Vrana NE. 2015. Macrophage responses to implants: prospects for personalized medicine. J Leukoc Biol. 98:953–962. doi:https://doi.org/10.1189/jlb.5VMR0415-166R
- Li Y, Liu X, Tang K, Wang P, Zeng Z, Guo Y, Wang X. 2019. Excisionase in Pf filamentous prophage controls lysis-lysogeny decision-making in Pseudomonas aeruginosa. Mol Microbiol. 111:495–513. doi:https://doi.org/10.1111/mmi.14170
- Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol. 196:2355–2366. doi:https://doi.org/10.1128/JB.01493-14
- Malachowa N, Kobayashi SD, Freedman B, Dorward DW, DeLeo FR. 2013. Staphylococcus aureus leukotoxin GH promotes formation of neutrophil extracellular traps. J Immunol. 191:6022–6029. doi:https://doi.org/10.4049/jimmunol.1301821
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 4:e5822. doi:https://doi.org/10.1371/journal.pone.0005822
- Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia. 169:323–331. doi:https://doi.org/10.1007/s11046-009-9264-y
- Mazzoleni V, Zimmermann K, Smirnova A, Tarassov I, Prévost G. 2021. Staphylococcus aureus Panton-Valentine leukocidin triggers an alternative NETosis process targeting mitochondria. FASEB J. 35:e21167. doi:https://doi.org/10.1096/fj.201902981R
- Meirelles LA, Newman DK. 2018. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol Microbiol. 110:995–1010. doi:https://doi.org/10.1111/mmi.14132
- Missiakas D, Winstel V. 2021. Selective host cell death by Staphylococcus aureus: a strategy for bacterial persistence. Front Immunol. 11:621733. doi:https://doi.org/10.3389/fimmu.2020.621733
- Mlynek KD, Bulock LL, Stone CJ, Curran LJ, Sadykov MR, Bayles KW, Brinsmade SR. 2020. Genetic and biochemical analysis of CodY-mediated cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J Bacteriol. 202:e00593–19. doi:https://doi.org/10.1128/JB.00593-19
- Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 14:255–261. doi:https://doi.org/10.1016/s0958-1669(03)00036-3
- Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011a. Extracellular DNA in biofilms. Int J Artif Organs. 34:824–831. doi:https://doi.org/10.5301/ijao.5000051
- Montanaro L, Speziale P, Campoccia D, Ravaioli S, Cangini I, Pietrocola G, Giannini S, Arciola CR. 2011b. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 6:1329–1349. doi:https://doi.org/10.2217/fmb.11.117
- Nagakubo T, Nomura N, Toyofuku M. 2020. Cracking open bacterial membrane vesicles. Front Microbiol. 10:3026. doi:https://doi.org/10.3389/fmicb.2019.03026
- Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 38:213–231. doi:https://doi.org/10.1046/j.1365-2958.2000.02135.x
- Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: oxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 182:4533–4544. doi:https://doi.org/10.1128/JB.182.16.4533-4544.2000
- Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 41:341–352. doi:https://doi.org/10.3109/1040841X.2013.841639
- Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol. 322:207–228. doi:https://doi.org/10.1007/978-3-540-75418-3_10
- Paganelli FL, Willems RJ, Jansen P, Hendrickx A, Zhang X, Bonten MJ, Leavis HL. 2013. Enterococcus faecium biofilm formation: identification of major autolysin AtlAEfm, associated Acm surface localization, and AtlAEfm-independent extracellular DNA Release. mBio. 4:e00154. doi:https://doi.org/10.1128/mBio.00154-13
- Papayannopoulos V. 2018. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 18:134–147. doi:https://doi.org/10.1038/nri.2017.105
- Pérez-Cruz C, Delgado L, López-Iglesias C, Mercade E. 2015. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One. 10:e0116896. doi:https://doi.org/10.1371/journal.pone.0116896
- Pestrak MJ, Chaney SB, Eggleston HC, Dellos-Nolan S, Dixit S, Mathew-Steiner SS, Roy S, Parsek MR, Sen CK, Wozniak DJ. 2018. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog. 14:e1006842. doi:https://doi.org/10.1371/journal.ppat.1006842
- Pires DP, Melo LDR, Azeredo J. 2021. Understanding the complex phage-host interactions in biofilm communities. Annu Rev Virol. 8:73–94. doi:https://doi.org/10.1146/annurev-virology-091919-074222
- Popp PF, Mascher T. 2019. Coordinated cell death in isogenic bacterial populations: sacrificing some for the benefit of many? J Mol Biol. 431:4656–4669. doi:https://doi.org/10.1016/j.jmb.2019.04.024
- Price CE, Brown DG, Limoli DH, Phelan VV, O’Toole GA. 2020. Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J Bacteriol. 202:e00559–19. doi:https://doi.org/10.1128/JB.00559-19
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology (Reading). 153:2083–2092. doi:https://doi.org/10.1099/mic.0.2007/006031-0
- Ravaioli S, Campoccia D, Speziale P, Pietrocola G, Zatorska B, Maso A, Presterl E, Montanaro L, Arciola CR. 2020. Various biofilm matrices of the emerging pathogen Staphylococcus lugdunensis: exopolysaccharides, proteins, eDNA and their correlation with biofilm mass. Biofouling. 36:86–100. doi:https://doi.org/10.1080/08927014.2020.1716217
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 104:8113–8118. doi:https://doi.org/10.1073/pnas.0610226104
- Ríos-López AL, González GM, Hernández-Bello R, Sánchez-González A. 2021. Avoiding the trap: mechanisms developed by pathogens to escape neutrophil extracellular traps. Microbiol Res. 243:126644. doi:https://doi.org/10.1016/j.micres.2020.126644
- Rose SJ, Babrak LM, Bermudez LE. 2015. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PLoS One. 10:e0128772. doi:https://doi.org/10.1371/journal.pone.0128772
- Salgado-Pabón W, Jain S, Turner N, van der Does C, Dillard JP. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol. 66:930–947. doi:https://doi.org/10.1111/j.1365-2958.2007.05966.x
- Salgado-Pabón W, Du Y, Hackett KT, Lyons KM, Arvidson CG, Dillard JP. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J Bacteriol. 192:1912–1920. doi:https://doi.org/10.1128/JB.01357-09
- Sanchez-Torres V, Maeda T, Wood TK. 2010. Global regulator H-NS and lipoprotein NlpI influence production of extracellular DNA in Escherichia coli. Biochem Biophys Res Commun. 401:197–202. doi:https://doi.org/10.1016/j.bbrc.2010.09.026
- Sarkar S. 2020. Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA. Appl Microbiol Biotechnol. 104:6549–6564. 1. doi:https://doi.org/10.1007/s00253-020-10687-9
- Saunders SH, Tse ECM, Yates MD, Otero FJ, Trammell SA, Stemp EDA, Barton JK, Tender LM, Newman DK. 2020. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell. 182:919–932.e19. doi:https://doi.org/10.1016/j.cell.2020.07.006
- Seifert HS. 2017. Haemophilus spills its guts to make a biofilm. Proc Natl Acad Sci U S A. 114:8444–8446. doi:https://doi.org/10.1073/pnas.1711077114
- Seitz P, Blokesch M. 2014. DNA transport across the outer and inner membranes of naturally transformable Vibrio cholerae is spatially but not temporally coupled. mBio. 5:e01409–14. doi:https://doi.org/10.1128/mBio.01409-14
- Selders GS, Fetz AE, Radic MZ, Bowlin GL. 2017. An overview of the role of neutrophils in innate immunity, inflammation and host-biomaterial integration. Regen Biomater. 4:55–68. doi:https://doi.org/10.1093/rb/rbw041
- Shanker E, Federle MJ. 2017. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes (Basel). 8:15. doi:https://doi.org/10.3390/genes8010015
- Shields RC, Mokhtar N, Ford M, Hall MJ, Burgess JG, ElBadawey MR, Jakubovics NS. 2013. Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS One. 8:e55339. doi:https://doi.org/10.1371/journal.pone.0055339
- Singh JA, Yu S, Chen L, Cleveland JD. 2019. Rates of total joint replacement in the United States: future projections to 2020-2040 using the National Inpatient Sample. J Rheumatol. 46:1134–1140. doi:https://doi.org/10.3899/jrheum.170990
- Soe YM, Bedoui S, Stinear TP, Hachani A. 2021. Intracellular Staphylococcus aureus and host cell death pathways. Cell Microbiol. 23:e13317. doi:https://doi.org/10.1111/cmi.13317
- Speziale P, Pietrocola G. 2020. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front Microbiol. 11:2054. doi:https://doi.org/10.3389/fmicb.2020.02054
- Steinberg BE, Grinstein S. 2007. Unconventional roles of the NADPH oxidase: signaling, ion homeostasis, and cell death. Sci Stke. 2007:pe11. doi:https://doi.org/10.1126/stke.3792007pe11
- Steinmoen H, Knutsen E, Håvarstein LS. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci U S A. 99:7681–7686. doi:https://doi.org/10.1073/pnas.112464599
- Takei H, Araki A, Watanabe H, Ichinose A, Sendo F. 1996. Rapid killing of human neutrophils by the potent activator phorbol 12-myristate 13-acetate (PMA) accompanied by changes different from typical apoptosis or necrosis. J Leukoc Biol. 59:229–240. doi:https://doi.org/10.1002/jlb.59.2.229
- Tamura M, Ajayi T, Allmond LR, Moriyama K, Wiener-Kronish JP, Sawa T. 2004. Lysophospholipase A activity of Pseudomonas aeruginosa type III secretory toxin ExoU. Biochem Biophys Res Commun. 316:323–331. doi:https://doi.org/10.1016/j.bbrc.2004.02.050
- Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, Uchiyama H, Nomura N. 2010. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl Environ Microbiol. 76:3732–3739. doi:https://doi.org/10.1128/AEM.02794-09
- Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 342:863–866. doi:https://doi.org/10.1126/science.1242255
- Thanabalasuriar A, Scott BNV, Peiseler M, Willson ME, Zeng Z, Warrener P, Keller AE, Surewaard BGJ, Dozier EA, Korhonen JT, et al. 2019. Neutrophil extracellular traps confine Pseudomonas aeruginosa ocular biofilms and restrict brain invasion. Cell Host Microbe. 25:526–536.e4. doi:https://doi.org/10.1016/j.chom.2019.02.007
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 72:1022–1036. doi:https://doi.org/10.1111/j.1365-2958.2009.06703.x
- Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 190:5690–5698. doi:https://doi.org/10.1128/JB.00314-08
- Toyofuku M. 2019. Bacterial communication through membrane vesicles. Biosci Biotechnol Biochem. 83:1599–1605. doi:https://doi.org/10.1080/09168451.2019.1608809
- Toyofuku M, Cárcamo-Oyarce G, Yamamoto T, Eisenstein F, Hsiao CC, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L. 2017. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun. 8:481. doi:https://doi.org/10.1038/s41467-017-00492-w
- Toyofuku M, Nomura N, Eberl L. 2019. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 17:13–24. doi:https://doi.org/10.1038/s41579-018-0112-2
- Tromp AT, van Strijp JAG. 2020. Studying staphylococcal leukocidins: a challenging endeavor. Front Microbiol. 11:611. doi:https://doi.org/10.3389/fmicb.2020.00611
- Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, et al. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun. 7:11220. doi:https://doi.org/10.1038/ncomms11220
- Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5:e1000639. doi:https://doi.org/10.1371/journal.ppat.1000639
- Van Gennip M, Christensen LD, Alhede M, Phipps R, Jensen PØ, Christophersen L, Pamp SJ, Moser C, Mikkelsen PJ, Koh AY, et al. 2009. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 117:537–546. doi:https://doi.org/10.1111/j.1600-0463.2009.02466.x
- Vandenesch F, Lina G, Henry T. 2012. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol. 2:12. doi:https://doi.org/10.3389/fcimb.2012.00012
- Vitse J, Devreese B. 2020. The contribution of membrane vesicles to bacterial pathogenicity in cystic fibrosis infections and healthcare associated pneumonia. Front Microbiol. 11:630. doi:https://doi.org/10.3389/fmicb.2020.00630
- von Eiff C, Arciola CR, Montanaro L, Becker K, Campoccia D. 2006. Emerging Staphylococcus species as new pathogens in implant infections. Int J Artif Organs. 29:360–367. doi:https://doi.org/10.1177/039139880602900405
- Wilton M, Charron-Mazenod L, Moore R, Lewenza S. 2015. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 60:544–553. doi:https://doi.org/10.1128/AAC.01650-15
- Wongkaewkhiaw S, Kanthawong S, Bolscher JGM, Nazmi K, Taweechaisupapong S, Krom BP. 2020. DNase-mediated eDNA removal enhances D-LL-31 activity against biofilms of bacteria isolated from chronic rhinosinusitis patients. Biofouling. 36:1117–1128. doi:https://doi.org/10.1080/08927014.2020.1857741
- Yousefi S, Stojkov D, Germic N, Simon D, Wang X, Benarafa C, Simon HU. 2019. Untangling “NETosis” from NETs. Eur J Immunol. 49:221–227. doi:https://doi.org/10.1002/eji.201747053
- Zafra O, Lamprecht-Grandío M, de Figueras CG, González-Pastor JE. 2012. Extracellular DNA release by undomesticated Bacillus subtilis is regulated by early competence. PLoS One. 7:e48716. doi:https://doi.org/10.1371/journal.pone.0048716
- Zatorska B, Arciola CR, Haffner N, Segagni Lusignani L, Presterl E, Diab-Elschahawi M. 2018. Bacterial extracellular DNA production is associated with outcome of prosthetic joint infections. Biomed Res Int. 2018:1067413. doi:https://doi.org/10.1155/2018/1067413
- Zawrotniak M, Rapala-Kozik M. 2013. Neutrophil extracellular traps (NETs) - formation and implications. Acta Biochim Pol. 60:277–284. doi:https://doi.org/10.18388/abp.2013_1983
- Zhao J, Wang Q, Li M, Heijstra BD, Wang S, Liang Q, Qi Q. 2013. Escherichia coli toxin gene hipA affects biofilm formation and DNA release. Microbiology (Reading). 159:633–640. doi:https://doi.org/10.1099/mic.0.063784-0
