Abstract
Background
Renal cell carcinoma (RCC), arising from the renal tubular epithelium, is one of the most common types of genitourinary malignancies. Based on the Gene Expression Omnibus (GEO) database (GSE100666), S100 calcium-binding protein A8 (S100A8) was highly expressed in RCC tissues. S100A8, an inflammatory regulatory factor, has emerged as an important mediator associated with the occurrence and development of cancer.
Materials and Methods
The Gene Expression Omnibus (GEO) database was used to identify the key genes and investigate the main signaling pathways in RCC. Human RCC samples and corresponding adjacent normal tissues were collected in our hospital. The expression of S100A8 in human RCC samples was detected using western blotting and immunohistochemical analysis. S100A8 overexpression or knockdown was mediated by using Lipofectamine 3000 in human renal cell carcinoma cell line 786-O and ACHN cells. Basic experiments, including MTT and cell apoptosis assays, were utilized for investigating the function of S100A8 in RCC. Furthermore, the levels of inflammation were also evaluated in 786-O and ACHN cells.
Results
In the current study, we found that downregulation of S100A8 inhibited proliferation and promoted apoptosis in 786-O and ACHN RCC cells. Of note, S100A8 silencing downregulated the phosphorylation of NF-κB p65, thereby decreasing the levels of TNF-α, cleaved caspase1, and MMP9. By contrast, S100A8 upregulation could increase these expressions.
Conclusion
Overall, S100A8 knockdown restrained RCC malignant biological properties, which was associated with the deactivation of the NF-κB signaling pathway. This present study demonstrates new insights that S100A8 may be a potential therapeutic target in RCC.
Introduction
Renal cell carcinoma (RCC) is one of the most aggressive genitourinary malignancies, with about 400, 000 new cases diagnosed and more than 180, 000 deaths per year worldwide [Citation1]. RCC progression is associated with both the cell-extrinsic and cell-intrinsic processes, which include excessive proliferation, inhibitory apoptosis, alteration of the stromal environment, etc. Although great advancements were achieved in the treatment of RCC over the past decades, multiple patients were subjected to disease progression after surgery or combined therapy [Citation2]. New therapeutic target identification is crucial to supply better treatment approaches for RCC.
As a new subject of biological research, bioinformatics was broadly considered as using statistics, informatics, and mathematics to analyze biological data [Citation3]. Besides, bioinformatics is a crucial element that analyze data from molecular genetic tests in clinical laboratories [Citation4]. It is important to predict interactions among differentially expressed genes (DEGs) to identify essential factors and investigate key signaling pathways in RCC.
S100 calcium-binding protein A8 (S100A8), as a regulator of the immune response and inflammatory process, is involved in multiple cancerous progressions [Citation5]. Elevated levels of S100A8 were observed in various cancers, such as gastric cancer [Citation6], prostate cancer [Citation7], and colorectal carcinoma [Citation8]. Moreover, S100A8 was associated with the malignant phenotype of cancer. Upregulation of S100A8 induced disseminative progression in lung cancer [Citation9]. S100A8 silencing revealed the reduction of anaplastic thyroid carcinoma tumorigenesis [Citation10]. Interestingly, the level of S100A8 was dramatically increased in carcinoma samples of patients with RCC. Upregulation of S100A8 was associated with poor prognosis [Citation11]. However, the mechanism of S100A8 on RCC progression is rarely known.
Inflammation is a key driver of many diseases. Even more important is the possibility that inflammatory insults are associated with malignancy [Citation12]. Nuclear factor kappaB (NF-κB) activation could promote tumorigenic autocrine signaling, and this was shown to be necessary on tumorigenesis. Inhibition of NF-κB and tumor necrosis factor-α (TNF-α) led to apoptosis of transformed hepatocytes and failure in hepatocellular tumor formation [Citation13]. Furthermore, caspase1 inactivation suppressed proliferation in non-small cell lung cancer cells, subsequently restraining the progression of carcinogenesis [Citation14]. Notably, S100A8 was reported to increase the secretion of the pro-inflammatory cytokine TNF-α and activate the NF-κB signaling pathway in pancreatic cancer cells, as well as promoting the aggressive nature of cancer [Citation15]. Nevertheless, little is known about the function of S100A8 in RCC progression.
Materials and methods
Identification of DEGs and bioinformatics analysis
The gene chip data GSE100666 searching from the GEO database website (https://www.ncbi.nlm.nih.gov/) was used as the data source of bioinformatics analysis. GSE100666 is based on the GPL16951 Platform (Phalanx Human OneArray Ver. 6 Release 1) and included 3 RCC tissues and 3 corresponding adjacent normal tissues. The GEO2R tool was used to analyze microarray data related to renal cell carcinoma. Moreover, functional annotations, including Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses of biological processes (BP), molecular function (MF), and cellular component (CC) were used to identify interactome pathways in RCC.
Patient’s specimens
Human RCC samples (Carcinoma) and corresponding adjacent normal tissues (Normal) were collected from the Hongqi Hospital Affiliated to Mudanjiang Medical University after written informed consent was obtained from the patients. The study was approved by the Ethics Committee of Hongqi Hospital Affiliated to Mudanjiang Medical University (Approval Number: 202023).
Immunohistochemical analysis
Immunohistochemistry for S100A8 was performed on 5 μm thickness paraffin-embedded tissue sections. After antigen retrieval and endogenous peroxidase blocking, the slides were incubated with S100A8 rabbit polyclonal antibody (Abclonal, China) at a dilution of 1: 100 at 4 °C overnight. Subsequently, the goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (IgG-HRP, ThermoFisher SCIENTIFIC, USA) was used to detect primary rabbit immunoglobulins. S100A8 staining was visualized with a diaminobenzidine substrate (MXB Biotechnologies, China).
Cell culture and transfection
Human renal cell carcinoma cell line 786-O and ACHN cells purchased from iCell Bioscience lnc (China) were routinely grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, USA) or Minimum Eagle’s medium (MEM, Solarbio, China) supplemented with 10% fetal bovine serum (FBS, Tianhang Biotechnology, China) at 37 °C with 5% CO2. Cell transfection was carried out using Lipofectamine 3000 reagent (Invitrogen, USA) with S100A8 plasmid or siRNA-S100A8. The vector and siRNA negative control (si-NC) were used as corresponding controls. At 48 h post-transfection, cells were harvested for the following experiments.
Cell proliferation
The transfected cells were seeded at a density of 4 × 103 cells per well in a 96-well plate. Cell proliferation was detected after 48 h by adding to each well 50 μl of MTT (BeyotimeBiotech, China) and incubating at 37 °C for 4 h. After that, the reaction was stopped by adding 150 μl of DMSO, and the absorbance was tested in a microplate reader (BioTek, USA) at 570 nm.
Cell apoptosis
Annexin V-FITC/PI Apoptosis Detection Kit (KeyGene Biotech., China) was used to access the function of S100A8 in cell apoptosis. After 48 h of transfection, the cells in each group were incubated with annexin-v-FITC (5 μl) and PI (5 μl) for 15 min, followed analyzing on flow cytometry (Agilent, USA).
RNA isolation and real-time PCR
Total RNA was extracted from transfected cells using TRIpure reagent (BioTeke Bio., China) and converted into cDNA using oligo (dT) primers and reverse transcriptases (BeyotimeBiotech). Subsequently, real-time PCR was conducted to determine the gene expression of S100A8, TNF-α, and MMP9. Primer sequences were listed in . The results were normalized to β-actin CT values and the ΔΔCT method to determine fold changes (2−ΔΔCt) was used to calculate mRNA expression.
Table 1. The nucleotide sequences of PCR primers.
Western blotting
RIPA lysis (Solarbio) was used to extract total proteins from the tumor samples of RCC patients and transfected cells. The concentration of the extracted protein was detected using BCA Protein Assay Kit (Solarbio). Equal amounts of protein were separated by 10% or 17% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at 80 v for 2.5 h and then transfer-embedded onto a polyvinylidene difluoride (PVDF) membrane at 80 v for 1.5 h. After washing with TBST and blocking with 5% skim milk, members were hybridized with specific antibodies: polyclonal rabbit anti-S100A8 (1: 400, Abclonal), polyclonal rabbit anti-p-NF-κB p65 (1: 400, Affinity Biosciences, China), polyclonal rabbit anti-TNF-α (1: 500, Affinity Biosciences), polyclonal rabbit anti-MMP9 (1: 400, Affinity Biosciences), and monoclonal rabbit anti-cleaved caspase1 (1: 1000, Cell Signaling Technology, USA) at 4 °C overnight. Blots were washed and incubated with goat anti-rabbit IgG-HRP (1: 5000, Solarbio) at 37 °C for 45 min. After washing, the blots were exposed with enhanced chemiluminescence regent. Protein expression was quantified by Gel-Pro-Analyzer and normalized to β-actin.
Statistical analysis
Data were analyzed using GraphPad 8.0 and were expressed as mean ± standard deviation (STDEV). One-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test was performed to identify differences among inter-groups, considering a significance with the P value < 0.05.
Results
Gene expression profiling based on the gene chip data
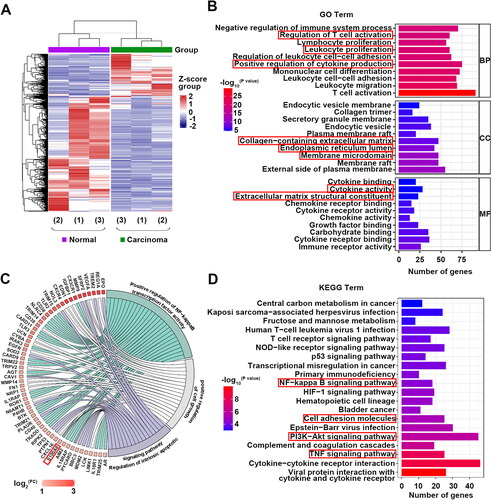
Gse100666 was analyzed online by GEO2R and 3831 DEGs (1071 up-regulated differentially expressed genes (DEGs) and 2760 down-regulated DEGs) were obtained and presented in a heatmap (|Log2(FC)|≥1.2, p < 0.05, ). Further, the top 10 GO annotations and top 20 KEGG pathway analyzes were shown in . Up-regulated DEGs were remarkably enriched in inflammatory biological processes, such as positive regulation of cytokine production. As for CC, the DEGs were enriched in the endoplasmic reticulum lumen and membrane microdomain. In addition, MF analysis showed that the DEGs were enriched in cytokine activity. KEGG pathway analysis revealed that the DEGs were relevant to the NF-κB signaling pathway (). Notably, enriched GO terms included positive regulation of cell growth, regulation of intrinsic apoptotic signaling pathway, and positive regulation of NF-κB transcription factor activity. These above processes were all associated with the up-regulated gene S100A8 (). This indicated that S100A8 alteration might be associated with the occurrence and development of RCC. Hence, the role of S100A8 in RCC was investigated in the following experiments.
Figure 1. Transcriptome profiling between RCC tumor tissues and corresponding adjacent normal tissues (GSE100666). DEGs were shown in the heatmaps (A). Top 10 GO analyses terms of BP, CC, and MF (B). GO analyses terms of BP that associated with cell growth, apoptosis, and inflammation (C). TOP 20 KEGG pathway enrichment analyses of DEGs (D).
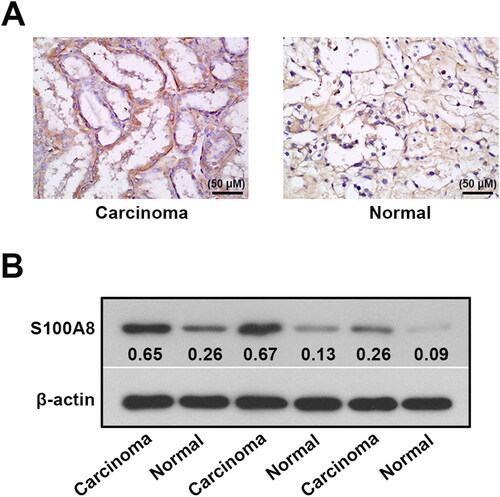
S100A8 expression was elevated in the renal cell carcinoma tissues of patients with RCC
Upregulation of S100A8 was observed in carcinoma samples compared to normal tissues using immunohistochemical staining (). An elevation of S100A8 in the carcinoma samples was also shown in the western blotting analysis and reinforced the hypothesis of S100A8 might act as an oncogene in RCC ().
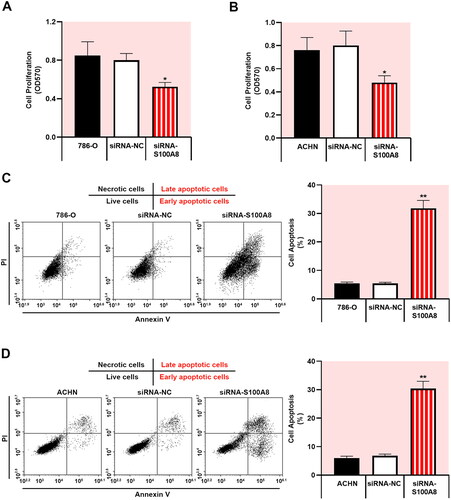
Knockdown of S100A8 inhibited RCC cell growth and promoted apoptosis
To evaluate the functional regulatory of S100A8 involved in RCC tumorigenesis, S100A8 was depleted in 786-O and ACHN cells using specific siRNA (Supplementary figure 1). S100A8 silencing significantly suppressed cell proliferation in RCC cells (, p < 0.05). Furthermore, depicted the image view of different stages of apoptosis of cells. Downregulation of S100A8 remarkably facilitated the apoptosis of 786-O and ACHN cells (p < 0.01). Therefore, our results indicated that S100A8 knockdown restrained cell proliferation and accelerated apoptosis in the regulation of RCC progression.
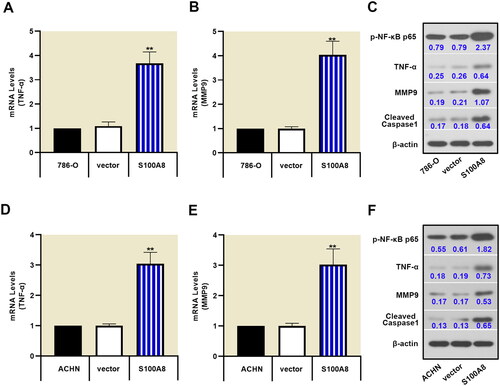
Silenced-S100A8 inactivated the NF-κB signaling pathway
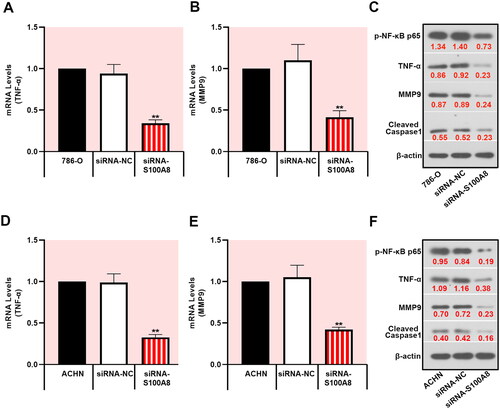
Several inflammatory biological processes were identified by transcriptome array analysis in RCC tissues and corresponding adjacent normal tissues. We chose to investigate the NF-κB signaling pathway, which causes the excessive proliferation of RCC cells [Citation16]. S100A8 knockdown decreased the phosphorylation of NF-κB, as well as inhibiting the expression of TNF-α, MMP9, and cleaved caspase-1 in 786-O and ACHN cells (). Additionally, overexpression of S100A8 promoted the NF-κB signaling pathway (Supplementary figure 2 and ). These data demonstrated that S100A8 might be essential to inflammation-induced cancer.
Figure 4. Downregulation of S100A8 suppressed the NF-κB signaling pathway. Real-time PCR was performed to access the mRNA expression of TNF-α (A, D) and MMP9 (B, E) in 786-O and ACHN cells, respectively. Protein expressions of p-NF-κB p65, TNF-α, MMP9, and cleaved caspase1 were determined by western blot (C, F).
Discussion
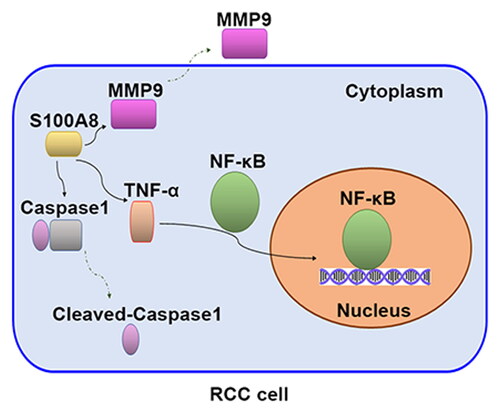
In this study, using GEO microarray data and clinical samples determination, we identified that S100A8 was upregulated in RCC tissues. Of note, S100A8 knockdown inhibited proliferation and promoted apoptosis in RCC cells. Mechanistically, the tumor-suppressive effect of silenced-S100A8 was associated with the inactivation of the NF-κB pathway (Graphical abstract). These results might point toward the S100A8 as a therapy target in RCC.
As multiple molecular factors were associated with RCC progression, there is a great possibility to identify the novel drug target. In the first part of this study, we analyzed the expression profile of genes between RCC samples and corresponding adjacent normal tissues. Our results showed that upregulated genes were remarkably enriched in the pathway of inflammation. Chronic inflammation may cause the occurrence and development of several diseases, including cancer [Citation17]. Based on the GO analysis, several DEGs were identified, such as fibronectin (FN), Tripartite motif (TRIM), S100A8, etc. FN1 belongs to the FN family, which is associated with epithelial-mesenchymal transition and promotes tumor cell invasion [Citation18]. TRIMs, associated with several forms of cancer, were clearly reported in RCC progression [Citation19]. As one of the DEGs connected with GO analysis involved in cell growth, apoptosis, and inflammation, S100A8 might be associated with the pathogenesis of RCC at the molecular level.
The S100 protein family, exclusively expressed in vertebrates, is reported to have a crucial role in regulating inflammation-related processes during multiple diseases, especially in malignancies [Citation20]. S100 proteins consist of 25 core paralogs, including S100A1-S100A18, S100B, S100G, etc [Citation21]. S100A7 was deemed to present as a diagnostic biomarker for oral squamous cell carcinoma [Citation22]. S100A9 was an important driver for hepatocellular carcinoma [Citation23]. Additionally, the expressions of S100A8, S100A9, and S100B were upregulated in the serum of glioma patients and might act as biomarkers for prognosis [Citation24]. In the present study, S100A8 expression was observably elevated in RCC samples compared with the normal tissues, not only by microarray analysis in GEO datasets but also by immunohistochemistry and western blot. Such consistent results indicated that S100A8 was associated with the development of RCC.
It is well known that excessive proliferation and reduced apoptosis are the main features of cancer. Identifying an attractive therapeutic biomarker for the treatment of RCC can be expected upon regulating proliferation and apoptosis. A recent study showed that downregulation of S100A8 suppressed cell growth in glioma cells [Citation25]. S100A8 silencing inhibited the proliferation of nasopharyngeal carcinoma cells via restraining the PI3K/Akt signaling pathway [Citation26]. Furthermore, S100A8 was associated with endoplasmic reticulum stress transmission that provoke tumor progression [Citation27]. Our findings indicated that blockade of S100A8 inhibited proliferation and promoted apoptosis in RCC cells. GO and KEGG analysis indicated that the DEGs were enriched in the endoplasmic reticulum lumen and relevant to the PI3K/Akt signaling pathway in RCC. Based on the above studies, S100A8 knockdown might be an effective therapeutic strategy for the treatment of RCC via suppressing the PI3K/Akt pathway and facilitating the endoplasmic reticulum-mediated apoptosis pathway. Whether these two signaling pathways were regulated by S100A8 for tumor progression needs further investigation.
Previous studies manifested that NF-κB regulates cell proliferation, migration, and chemotherapy resistance in various cancers [Citation28]. As an essential transcription factor, NF-κB was identified as a crucial player in osteosarcoma progression [Citation29]. Inactivation of the NF-κB signaling pathway was concurrent with inhibitory effects on cell migration in triple-negative breast cancer cells [Citation30]. Inhibition of the NF-κB/Nod-like receptor protein 3 (NLRP3) pathway was associated with hepatocellular carcinoma cell apoptosis [Citation31]. NLRP3 is a multiprotein complex, which was shown to be involved in caspase1 activation, interleukin-1β (IL-1β) and TNF-α release, and pyroptotic cell death (pyroptosis) promotion [Citation32,Citation33]. Reduced tumor growth and induction of robust anti-cancer immunity were exhibited in NLRP3-deficient mice [Citation34]. Importantly, S100A8 promoted cholangiocarcinoma metastasis via activating the NF-κB signaling pathway [Citation35]. S100A8 driven NLRP3 inflammasome-mediated IL-1β was dependent upon NF-κB activation [Citation36]. In line with the previous studies, downregulation of S100A8 inhibited NF-κB p65 activation, subsequently decreasing the levels of TNF-α, cleaved caspase1, and MMP9 in RCC cells. Besides, S100A8 overexpression promoted these expressions. As a well-known metastatic inducer, MMP9 can cleave extracellular matrix (ECM) proteins to adjust ECM remodeling, which is closely connected with cancer progression [Citation37]. Upregulation of S100A8 notably facilitated the expression of MMP9, which was associated with nasopharyngeal carcinoma cell invasion and metastasis [Citation38]. NLRP3-caspase1 inflammasome was reported to directly activate MMP9 [Citation39]. Taking into consideration that S100A8 are implicated in the development of various cancers, S100A8 might activate the NF-κB/NLRP3 signaling pathway that led to RCC progression.
In conclusion, our observations illuminated that S100A8 was elevated in the RCC samples. Downregulation of S100A8 suppressed cell proliferation and facilitated apoptosis in RCC cells with the mechanism involving the inactivation of the NF-κB signaling pathway. These results had significant implications and indicated that S100A8 might be a key factor in RCC progression.
Supplemental Material
Download Zip (1,002.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets partly generated in this study are publicly available in GEO at GSE100666 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100666]. All data generated during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Makino T, Kadomoto S, Izumi K, Mizokami A. Epidemiology and prevention of renal cell carcinoma. Cancers (Basel). 2022;14(16):4059. doi:https://doi.org/10.3390/cancers14164059.
- Goebell PJ, Staehler M, Muller L, et al. Changes in treatment reality and survival of patients with advanced clear cell renal cell carcinoma - analyses from the german clinical RCC-registry. Clin Genitourin Cancer. 2018;16(6):1–9. doi:https://doi.org/10.1016/j.clgc.2018.06.006.
- Hephzibah Cathryn R, Udhaya Kumar S, Younes S, Zayed H, George Priya Doss C. A review of bioinformatics tools and web servers in different microarray platforms used in cancer research. Adv Protein Chem Struct Biol. 2022;131:85–164. doi:https://doi.org/10.1016/bs.apcsb.2022.05.002.
- Oliver GR, Hart SN, Klee EW. Bioinformatics for clinical next generation sequencing. Clin Chem. 2015;61(1):124–135. doi:https://doi.org/10.1373/clinchem.2014.224360.
- Gebhardt C, Nemeth J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72(11):1622–1631. doi:https://doi.org/10.1016/j.bcp.2006.05.017.
- Yong HY, Moon A. Roles of calcium-binding proteins, S100A8 and S100A9, in invasive phenotype of human gastric cancer cells. Arch Pharm Res. 2007;30(1):75–81. doi:https://doi.org/10.1007/BF02977781.
- Hermani A, De Servi B, Medunjanin S, Tessier PA, Mayer D. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res. 2006;312(2):184–197. doi:https://doi.org/10.1016/j.yexcr.2005.10.013.
- Stulik J, Osterreicher J, Koupilova K, et al. The analysis of S100A9 and S100A8 expression in matched sets of macroscopically normal colon mucosa and colorectal carcinoma: the S100A9 and S100A8 positive cells underlie and invade tumor mass. Electrophoresis. 1999;20(4–5):1047–1054. doi:https://doi.org/10.1002/(SICI)1522-2683(19990101)20:4/5 < 1047::AID-ELPS1047 > 3.0.CO;2-E.
- Sumardika IW, Chen Y, Tomonobu N, et al. Neuroplastin-beta mediates S100A8/A9-induced lung cancer disseminative progression. Mol Carcinog. 2019;58(6):980–995. doi:https://doi.org/10.1002/mc.22987.
- Reeb AN, Li W, Sewell W, et al. S100A8 is a novel therapeutic target for anaplastic thyroid carcinoma. J Clin Endocrinol Metab. 2015;100(2):E232–42. doi:https://doi.org/10.1210/jc.2014-2988.
- An HJ, Koh HM, Song DH. S100A8 expression may have a prognostic value in CCRCC reflecting TNM staging and fuhrman nuclear grade. Anticancer Res. 2019;39(9):4681–4685. doi:https://doi.org/10.21873/anticanres.13650.
- Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi:https://doi.org/10.1038/nrc3611.
- Pikarsky E, Porat RM, Stein I, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–466. doi:https://doi.org/10.1038/nature02924.
- Zou J, Yang Y, Yang Y, Liu X. Polydatin suppresses proliferation and metastasis of non-small cell lung cancer cells by inhibiting NLRP3 inflammasome activation via NF-kappaB pathway. Biomed Pharmacother. 2018;108:130–136. doi:https://doi.org/10.1016/j.biopha.2018.09.051.
- Nedjadi T, Evans A, Sheikh A, et al. S100A8 and S100A9 proteins form part of a paracrine feedback loop between pancreatic cancer cells and monocytes. BMC Cancer. 2018;18(1):1255. doi:https://doi.org/10.1186/s12885-018-5161-4.
- Kumar A, Kumari N, Gupta V, Prasad R. Renal cell carcinoma: Molecular aspects. Indian J Clin Biochem. 2018;33(3):246–254. doi:https://doi.org/10.1007/s12291-017-0713-y.
- Shah A, Patel C. A concise review of inflammatory biomarkers targeted cancer therapy. Folia Med (Plovdiv). 2022;64(4):572–580. doi:https://doi.org/10.3897/folmed.64.e68365.
- Park J, Schwarzbauer JE. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2014;33(13):1649–1657. doi:https://doi.org/10.1038/onc.2013.118.
- Shen J, Wang R, Chen Y, et al. Comprehensive analysis of expression profiles and prognosis of TRIM genes in human kidney clear cell carcinoma. Aging (Albany NY). 2022;14(10):4606–4617. doi:https://doi.org/10.18632/aging.204102.
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15(2):96–109. doi:https://doi.org/10.1038/nrc3893.
- Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396(2):201–214. doi:https://doi.org/10.1042/BJ20060195.
- Sood A, Mishra D, Kharbanda OP, et al. Role of S100 A7 as a diagnostic biomarker in oral potentially malignant disorders and oral cancer. J Oral Maxillofac Pathol. 2022;26(2):166–172. doi:https://doi.org/10.4103/jomfp.jomfp_402_20.
- Zhong C, Niu Y, Liu W, et al. S100A9 Derived from chemoembolization-induced hypoxia governs mitochondrial function in hepatocellular carcinoma progression. Adv Sci (Weinh). 2022;9(30):e2202206. doi:https://doi.org/10.1002/advs.202202206.
- Wang H, Mao X, Ye L, Cheng H, Dai X. The role of the S100 protein family in glioma. J Cancer. 2022;13(10):3022–3030. doi:https://doi.org/10.7150/jca.73365.
- Xiong J, Wang T, Tang H, Lv Z, Liang P. Circular RNA circMAN2B2 facilitates glioma progression by regulating the miR-1205/S100A8 axis. J Cell Physiol. 2019;234(12):22996–23004. doi:https://doi.org/10.1002/jcp.28860.
- Wen L, Ding Y, Chen X, et al. Influences of S100A8 and S100A9 on proliferation of nasopharyngeal carcinoma cells through PI3K/Akt signaling pathway. Biomed Res Int. 2021;2021:9917365. doi:https://doi.org/10.1155/2021/9917365.
- Wei W, Zhang Y, Song Q, et al. Transmissible ER stress between macrophages and tumor cells configures tumor microenvironment. Cell Mol Life Sci. 2022;79(8):403. doi:https://doi.org/10.1007/s00018-022-04413-z.
- Pikarsky E, Ben-Neriah Y. NF-kappaB inhibition: a double-edged sword in cancer? Eur J Cancer. 2006;42(6):779–784. doi:https://doi.org/10.1016/j.ejca.2006.01.011.
- Li CH, Ku MC, Lee KC, et al. Magnolol suppresses ERK/NF-kappaB signaling and triggers apoptosis through extrinsic/intrinsic pathways in osteosarcoma. Anticancer Res. 2022;42(9):4403–4410. doi:https://doi.org/10.21873/anticanres.15940.
- Messeha SS, Zarmouh NO, Antonie L, Soliman KFA. Sanguinarine inhibition of TNF-alpha-induced CCL2, IKBKE/NF-kappaB/ERK1/2 signaling pathway, and cell migration in human triple-negative breast cancer cells. Int J Mol Sci. 2022;23(15):8329. doi:https://doi.org/10.3390/ijms23158329.
- Saber S, Ghanim AMH, El-Ahwany E, El-Kader EMA. Novel complementary antitumour effects of celastrol and metformin by targeting IkappaBkappaB, apoptosis and NLRP3 inflammasome activation in diethylnitrosamine-induced murine hepatocarcinogenesis. Cancer Chemother Pharmacol. 2020;85(2):331–343. doi:https://doi.org/10.1007/s00280-020-04033-z.
- Aghelan Z, Karima S, Khazaie H, et al. Interleukin-1alpha and tumor necrosis factor alpha as an inducer for reactive-oxygen-species-mediated NOD-like receptor protein 1/NOD-like receptor protein 3 inflammasome activation in mononuclear blood cells from individuals with chronic insomnia disorder. Euro J of Neurology. 2022;29(12):3647–3657. doi:https://doi.org/10.1111/ene.15540.
- Paik S, Kim JK, Silwal P, Sasakawa C, Jo EK. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell Mol Immunol. 2021;18(5):1141–1160. doi:https://doi.org/10.1038/s41423-021-00670-3.
- Papafragkos I, Grigoriou M, Boon L, Kloetgen A, Hatzioannou A, Verginis P. Ablation of NLRP3 inflammasome rewires MDSC function and promotes tumor regression. Front Immunol. 2022;13:889075. doi:https://doi.org/10.3389/fimmu.2022.889075.
- Pan S, Hu Y, Hu M, et al. S100A8 facilitates cholangiocarcinoma metastasis via upregulation of VEGF through TLR4/NF‑kappaB pathway activation. Int J Oncol. 2020;56(1):101–112. doi:https://doi.org/10.3892/ijo.2019.4907.
- Silveira AAA, Mahon OR, Cunningham CC, et al. S100A8 acts as an autocrine priming signal for heme-induced human Mvarphi pro-inflammatory responses in hemolytic inflammation. J Leukoc Biol. 2019;106(1):35–43. doi:https://doi.org/10.1002/JLB.3MIA1118-418RR.
- Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent advances. Sensors (Basel). 2018;18(10):3249. doi:https://doi.org/10.3390/s18103249.
- Hu W, Tao Z, Zhou Q, et al. Effects of S100 calcium-binding protein A8 (S100A8) and S100 calcium-binding protein A9 (S100A9) on matrix metalloproteinase (MMP) expression in nasopharyngeal carcinoma CNE-2 cells. Transl Cancer Res. 2021;10(4):1874–1884. doi:https://doi.org/10.21037/tcr-21-441.
- Ren P, Wu D, Appel R, et al. Targeting the NLRP3 inflammasome with inhibitor MCC950 prevents aortic aneurysms and dissections in mice. J Am Heart Assoc. 2020;9(7):e014044. doi:https://doi.org/10.1161/JAHA.119.014044.