 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Introduction: Particulate matter (PM) has various systemic effects. We researched the effects of PM on lung epithelial cells with next generation sequencing (NGS) and validated this with quantitative real-time polymerase chain reaction (qRT-PCR).
Methods: We cultured the group exposed to PM10 (Particulate matter less than 10 μm)-like fine dust (ERM® CZ120 fine dust) at a concentration of 50 μg/mL and the untreated group for seven days in one normal lung epithelial cell line (BEAS-2B) and four lung cancer epithelial cell lines (NCI-H358, HCC-827, A549, NCI-H292). Then, we extracted the RNA from the sample and performed NGS. As a result of NGS, various gene expressions were upregulated or downregulated. Among them, we selected the gene whose mean fold change was more than doubled and changed in the same direction in all five cell lines. Based on these genes, we selected the top 10 genes, either upregulated or downregulated, to validate with the qRT-PCR.
Results: There were the four genes that matched the NGS and qRT-PCR results, all of which were upregulated genes. The four genes are CYP1A1, CYP1B1, LINC01816, and BPIFA2. All four genes that matched the two results were upregulated genes and none of the downregulated genes matched.
Conclusion: CYP1A1 and CYP1B1 are known to cause lung cancer by metabolizing polycyclic aromatic hydrocarbons, and long noncoding RNA is also known to play an important role in lung cancer. Considering this, we thought PM10 might be associated with lung cancer by activating CYP1A1, CYP1B1, and LINC01816.
Introduction
South Korea’s air quality is one of the worst in the world. According to the 2018 Environmental Performance Index (EPI), compiled by Yale University and Columbia University in collaboration with the World Economic Forum, South Korea’s air quality ranked 119th among 180 countries. In detail the population-weighted average ambient concentration of PM2.5 (Particulate matter less than 2.5 μm) placed South Korea at 174 out of 180 countries (Wendling et al. Citation2018). In addition, according to The Economic consequences of outdoor air pollution that the Organization for Economic Cooperation and Development (OECD) released in 2016, they estimated that Korea’s premature mortality rate from air pollution will increase, with South Korea having the highest mortality rate among OECD countries by 2060 (OECD Citation2016). The PM10 is gradually decreasing in Seoul, South Korea, but it is still higher than in some other cities in OECD countries, including New York and London (Ministry of Environment Citation2016). Because of the influence of external inflows from China, the concentration of PM in the air is expected to remain high for several decades.
The composition of fine dust varies from region to region (Lee et al. Citation2018), and even in the same area, the composition of fine dust varies depending on the surrounding conditions, such as traffic volume, industrial emissions or the presence of incinerators. DNA damage can vary depending on the composition of fine dust. Higher traffic emissions can cause greater toxicity (Gerlofs-Nijland et al. Citation2007; Sharma et al. Citation2007).
It has been reported that PM has systemic consequences not only respiratory and cardiovascular diseases, but also metabolic disease, neurodegenerative disease, and premature birth (Thurston et al. Citation2017). Many studies report a high respiratory and cardiovascular mortality rate with exposure to PM (Halonen et al. Citation2009; Guaita et al. Citation2011; Perez et al. Citation2012). Lowering exposure to PM10 can reduce lung function decline, and this effect is more pronounced in tests that detect small airway function (Downs et al. Citation2007). Air pollution is associated with an increased risk of chronic obstructive pulmonary disease hospitalization and mortality (Gan et al. Citation2013). PM has been reported to increase the incidence, hospitalization, and emergency room visits because of asthma (Tecer et al. Citation2008; Guarnieri and Balmes Citation2014). Both PM10 and PM2.5 contribute to lung cancer incidence (Raaschou-Nielsen et al. Citation2013).
When assessing air pollution, three things should be considered: emissions, concentrations, and human risks. Although the analysis of the emissions and concentrations has been a subject of considerable study, there is currently insufficient research on the biological mechanisms of human risks, except for epidemiology. We are aware of the seriousness of air pollution, but we are not sure which mechanism is causing the problem. So, our team studied what changes in gene expression occur when PM10 is introduced into lung epithelial cells using next generation sequencing (NGS) and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR).
Methods
Cell culture and PM10 treatment
In this experiment, we used a total of five cell lines: BEAS-2B (human lung, bronchus normal cell), HCC-827 (human non-small cell lung cancer), NCI-H358 (human bronchiolar carcinoma), NCI-H292 (human lung carcinoma), and A549 (human lung carcinoma). We used lung epithelial cells with various characteristics for common results. BEAS-2B cells are normal lung epithelial cells, HCC-827 cells are cancer cells with EGFR mutations, NCI-H358 and NCI-H292 are epithelial like cancer cells, and A549 cells are similar to mesenchymal like cancer cells (Thomson et al. Citation2008). We purchased the NCI-H358 and HCC-827 from the Korea Cell Line Bank, and we purchased the A549, NCI-H292, and BEAS-2B from the American Type Culture Collection. We cultured the NCI-H358, NCI-H292, HCC-827, and the A549 cells from Roswell Park Memorial Institute 1640 and BEAS-2B in a keratinocyte serum free medium. We chose ERM® CZ120 fine dust as the PM10 because it is easy to use and it is used in the calibration of fine dust measuring devices in Korea and the particle size is also guaranteed. The ERM® CZ120 fine dust is mainly composed of arsenic, cadmium, lead, nickel, and other additional ingredients: aluminum, molybdenum, antimony, neodymium, barium, potassium, bromine, rubidium, cesium, samarium, calcium, scandium, cerium, silicon, chlorine, sodium, chromium, strontium, cobalt, tantalum, copper, terbium, dysprosium, thorium, elemental carbon, titanium, europium, total carbon, gallium, total organic carbon, gold, tungsten, hafnium, uranium, iron, vanadium, lanthanum, ytterbium, magnesium, zinc, manganese, and zirconium.
We diluted the PM10 in phosphate buffered saline to make a stock at a concentration of 10 μg/μL and stored them frozen. After stabilization, we seeded 3× NCI-H358, HCC-827, A549, NCI-H292, and the BEAS-2B cells in a 100PI dish for subculture. After 24 h, we exposed the PM10 to a final concentration of 50 μg/mL and cultured it for seven days. If the cell’s confluence was more than 80% during cultivation, we exposed the PM in the same manner following subculture. The control group was cultured for seven days without treating the PM10. We extracted RNA from the control group and PM treatment group and stored them at −80 °C.
Next generation sequencing
After extracting the total RNA from the samples, we enriched the mRNA of the eukaryotes by using Oligo(dT) magnetic beads. Adding the fragmentation buffer, the mRNA was interrupted to form short fragments (about 200 bp). Then we synthesized the first strand cDNA by random hexamer-primer using the mRNA fragments as templates. We added buffer, dNTPs, RNase H and DNA polymerase I to synthesize the second strand. We purified the double strand cDNA with the QIAquick PCR extraction kit and washed with elution buffer for end repair and poly(A) addition. Finally, we ligated sequencing adaptors to the fragments. We purified the required fragments with agarose gel electrophoresis and enriched them with PCR amplification. The library products were ready for sequencing analysis via the Illumina HiSeq™ 4000.
Quantitative real-time polymerase chain reaction
Reverse transcription
We mixed 1 μg of RNA extract with AccuPower® RocketScript™ Cycle, primer (Oligo dT 20mer) and dextrose water to make a 20 μL solution. We performed reverse transcription with 20 μL per reaction using a MyGenie™ 96 Gradient Thermal Block, under the following temperature profile: 12 cycles each consisting of 30 s at 37 °C, 4 min at 48 °C and 30 s at 55 °C, and one cycle of 10 min at 95 °C.
Real time-polymerase chain reaction (RT-PCR)
We performed RT-PCR tests using the AccuPower® 2X GreenStar Master Mix with the Exicycler™ 96 Real-Time Quantitative Thermal Block under the following temperature profile: one cycle of 10 min at 95 °C, 40 cycles of 5 s at 95 °C, 25 s at 58 °C and 30 s at 72 °C, and an extension at 65 °C for 5 min. We did an analysis of the melting curve at 65 °C ∼95 °C (1 °C/s).
lists the gene primers sequences.
Table 1. qRT-PCR primer information.
Results
Gene expression profiling by NGS
Based on the NGS results, we selected genes whose mean fold changes were more than doubled and which had been changed in the same direction in all five cell lines. We allowed up to two cell lines having no differences between the control and experimental groups per gene.
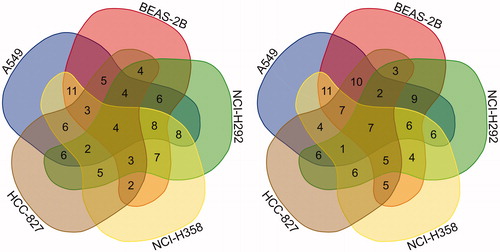
shows Venn diagrams of the twofold gene changes based on selected genes. The left Venn diagram shows the number of upregulated genes, and the right one shows the number of down regulated genes. The number of selected upregulated genes was 84 in total. Among them the number of genes upregulated in all five cell lines was four, and the number of genes upregulated in four cell lines was 20. The number of genes upregulated in three cell lines was 60. The number of selected down regulated genes was 92 in total. Among them, the number of downregulated genes in all 5 cell lines was 7, and the number of genes down regulated in 4 cell lines was 21. The number of genes down regulated in 3 cell lines was 64.
Figure 1. Venn diagram: the number of selected genes (Left: upregulated genes, Right: down regulated genes).
and show information on the top 10 of the selected genes that were upregulated and down regulated on NGS. Looking at the table, the values of fold change (
FC) for each cell line and the value of mean
FC are shown. The order of genes in the table was arranged in the order of the mean
FC.
Table 2. Top 10 upregulated genes based on NGS results.
Table 3. Top 10 down regulated genes based on NGS results.
Validation by qRT-PCR
Among the selected genes, the top 10 genes that were changed upward and downward were validated by qRT-PCR. and show the results of qRT-PCR. In the qRT-PCR results, there were some cases where RNA was not detected. In these cases, the fold change value was indicated as being not calculated (NC) and the mean fold change was calculated as being the fold change value of the remaining cell lines except for the cell line where RNA was not detected. When interpreting the results, cell lines whose fold change is NC were excluded. As in the case of NGS, we selected genes whose mean fold changes were more than doubled and which had been changed in the same direction in all cell lines on qRT-PCR results. Among the selected genes, those genes having three or more NC values were finally excluded. Consequently, four of the upregulated genes were identical to the NGS results: CYP1A1, LINC01816, BPIFA2, CYP1B1.
Table 4. Validation of the top 10 upregulated genes by qRT-PCR.
Table 5. Validation of the top 10 down regulated genes by qRT-PCR.
Discussion
PM is an air pollutant in which solid particles and liquid particles are mixed. The composition of PM varies depending upon the location, but generally it contains sulfates, nitrates, ammonium, other inorganic ions(sodium, potassium, calcium, magnesium and chloride), organic and elemental carbon, crustal material, particle-bound water, metals (cadmium, copper, nickel, vanadium and zinc), polycyclic aromatic hydrocarbons and biological components(allergens and microbial compounds) (WHO Citation2013). PM is classified by diameter. A PM10 is a particle having a diameter of less than 10 µm, and a PM2.5 is a particle having a diameter of less than 2.5 µm. The exposure of PM depends on how people breathe, but the particle size is directly related to the main causes of health problems (Brown et al. Citation2013). PM10 tends to settle in the upper airway and either the trachea or the bronchi because it settles rapidly (Atkinson et al. Citation2010).
It is already known that PM is a carcinogen, as stated in the publications of the World Health Organization (WHO) and the International Agency for Research on Cancer (IARC Citation2013) According to the ESCAPE study, the hazard ratio of lung cancer is 1.22 at a concentration of 10 μm/m3 in PM10 and 1.8 at a concentration of 5 μm/m3 in PM2.5 (Raaschou-Nielsen et al. Citation2013). PM has been found to have a greater effect on lung cancer incidence in smokers than in healthy people (Pope III Citation2002). In addition to lung cancer, PM has been reported to cause airway inflammation, increased inflammatory cytokines, and polymorphonuclear leukocytes on bronchoalveolar lavage or sputum, as well as an increased level of lung injury blood markers, such as 8-isoprostane and club cell secretory protein (Thurston et al. Citation2017).
Among the top 10 genes upregulated and downregulated genes in our study, those whose NGS and qRT-PCR results match are the CYP1A1, CYP1B1, LINC01816, and BPIFA2, as mentioned above. CYP1A1 and CYP1B1 encode a member of the cytochrome P450 superfamily of enzymes. The expression of CYP1A1 is induced by some polycyclic aromatic hydrocarbons (PAHs) or tobacco smoke, and they can metabolize some PAHs to carcinogenic intermediates (Shimada and Fujii-Kuriyama Citation2004). The expression can be regulated by heavy metal (Korashy and El-Kadi Citation2005; Anwar-Mohamed et al. Citation2009). There are many reports that CYP1A1 is linked to lung cancer. CYP1A1 can initiate carcinogenesis in lung cells through PAH metabolism (Uppstad et al. Citation2010). CYP1A1 plays an important role in lung DNA adduct formation and causes a higher susceptibility to lung cancer among women than men (Mollerup et al. Citation2006). Przygodzki et al. (Citation1998) reported that CYP1A1 activation contributes to lung cancer via p53 inactivation. Some studies show that CYP1A1 polymorphism contributes to lung cancer susceptibility (Taioli et al. Citation2003; Ji et al. Citation2012).
CYP1B1 metabolizes many endogenous and exogenous substrates such as PAH, cholesterol, steroids, and estrogens. And it activates procarcinogens to become carcinogens (Aklillu et al. Citation2005; Sissung et al. Citation2006; Gajjar et al. Citation2012). Piotr Sawrycki et al. (Citation2018) reported a definite link between CYP1B1 and lung cancer in smoker. Based on this, we thought that PM10 might be associated with lung cancer by activating the CYP1A1 and CYP1B1 genes.
BPIFA2 encodes a member of the PLUNC protein family (PLUNC – palate, lung, and nasal epithelium clone). BPIFA2 is known as a short PLUNC-2 (SPLUNC2). It is present in a gene cluster on chromosome 20 (Bingle and Bingle Citation2000; Bingle and Craven Citation2002; Bingle et al. Citation2011). Members of this family have been proposed to play a role in local antibacterial responses in the nose, mouth and upper respiratory pathway (Geetha et al. Citation2003; Gorr et al. Citation2011). Vladimir Prokopovic et al. (Citation2014) reported that BPIFA2 protein has an antibacterial effect. We thought that BPIFA2 was upregulated to protect the body from the inflammatory response of PM10.
LINC01816 is known as long non-coding RNA (lncRNA). The mutation and misregulation of lncRNAs are known to play an important role in cancer (Wapinski and Chang Citation2011; Yan et al. Citation2015). In addition, lncRNA has a strong potential for being a new biomarker and therapeutic target because of its genome-wide expression pattern and tissue-specific expression characteristics in various tissues (Bhan et al. Citation2017). So further study of LINC01816 is needed .
Most studies for gene expression of cells exposed to fine dust were analyzed through microarray or qRT-PCR and by using particle samples collected directly. These studies mainly used single cell line and exposed to PM2.5 or particles of undetermined size for less than 24 h (Choi et al. Citation2011; Gualtieri et al. Citation2012; Sun et al. Citation2012; Ding et al. Citation2014; Chang et al. Citation2016). Our study differs in that we used various cell lines for common results, exposed to PM10 for seven days, and analyzed through NGS. Studies using NGS for gene expression in this field are scanty. Our study has the strength of using NGS in gene expression.
This study is limited because it was an in vitro study. In addition, we used PM10 for the study, and there is the limitation that PM10 is mainly settles in the upper airway. However, other epidemiological studies show the link between PM10 and lung disease(Downs et al. Citation2007; Cadelis et al. Citation2014), so that would not be a big limitation. Rather, it will be a strength because many studies have been done with PM2.5. We used commercial particles as PM10 in this study. Therefore, it may be difficult to apply our results in areas with significantly different fine dust components. Lastly, we analyzed the genes that had matching NGS and qRT-PCR results because we thought the matched genes are greatly linked to health problems. However, since each test has many variables, it cannot be concluded that the unmatched genes are meaningless. Therefore, further studies of these genes are needed.
In conclusion PM10 appears to activate some genes, such as the CYP1A1, CYP1B1 and LINC01816. These genes seem to be related to lung cancer.
Author contributions
Dong Ho Park: writing, conceptualization, reviewing and editing; Daeun Kang: writing, methodology, reviewing and editing; In Beom Jung: writing, reviewing and editing, funding acquisition; Su Yel Lee: methodology, investigation; Se Jin Park: visualization, data management; Sun Jung Kwon: data management, reviewing; Ji Woong Son: conceptualization, reviewing and editing, funding acquisition, supervision.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Aklillu E, Øvrebø S, Botnen IV, Otter C, Ingelman-Sundberg M. 2005. Characterization of common CYP1B1 variants with different capacity for benzo[a]pyrene-7,8-dihydrodiol epoxide formation from benzo[a]pyrene. Cancer Res. 65:5105.
- Anwar-Mohamed A, Elbekai RH, El-Kadi AO. 2009. Regulation of CYP1A1 by heavy metals and consequences for drug metabolism. Expert Opin Drug Metab Toxicol. 5:501–521.
- Atkinson RW, Fuller GW, Anderson HR, Harrison RM, Armstrong B. 2010. Urban ambient particle metrics and health: a time-series analysis. Epidemiology. 21:501–511.
- Bhan A, Soleimani M, Mandal SS. 2017. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77:3965–3981.
- Bingle CD, Bingle L. 2000. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta. 1493:363–367.
- Bingle CD, Craven CJ. 2002. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum Mol Genet. 11:937–943.
- Bingle CD, Bingle L, Craven CJ. 2011. Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/PLUNC protein family. Biochem Soc Trans. 39:961–965.
- Brown JS, Gordon T, Price O, Asgharian B. 2013. Thoracic and respirable particle definitions for human health risk assessment. Part Fibre Toxicol. 10:12.
- Cadelis G, Tourres R, Molinie J. 2014. Short-term effects of the particulate pollutants contained in Saharan dust on the visits of children to the emergency department due to asthmatic conditions in Guadeloupe (French Archipelago of the Caribbean). PLoS One. 9:e91136.
- Chang J, Go YY, Park MK, Chae SW, Lee SH, Song JJ. 2016. Asian sand dust enhances the inflammatory response and mucin gene expression in the middle ear. Clin Exp Otorhinolaryngol. 9:198–205.
- Choi H, Shin DW, Kim W, Doh SJ, Lee SH, Noh M. 2011. Asian dust storm particles induce a broad toxicological transcriptional program in human epidermal keratinocytes. Toxicol Lett. 200:92–99.
- Ding X, Wang M, Chu H, Chu M, Na T, Wen Y, Wu D, Han B, Bai Z, Chen W, et al. 2014. Global gene expression profiling of human bronchial epithelial cells exposed to airborne fine particulate matter collected from Wuhan. Toxicol Lett. 228:25–33.
- Downs SH, Schindler C, Liu LJS, Keidel D, Bayer-Oglesby L, Brutsche MH, Gerbase MW, Keller R, Künzli N, Leuenberger P, et al.; SAPALDIA Team. 2007. Reduced exposure to PM10 and attenuated age-related decline in lung function. N Engl J Med. 357:2338–2347.
- Gajjar K, Martin-Hirsch PL, Martin FL. 2012. CYP1B1 and hormone-induced cancer. Cancer Lett. 324:13–30.
- Gan WQ, FitzGerald JM, Carlsten C, Sadatsafavi M, Brauer M. 2013. Associations of ambient air pollution with chronic obstructive pulmonary disease hospitalization and mortality. Am J Respir Crit Care Med. 187:721–727.
- Geetha C, Venkatesh SG, Dunn BHF, Gorr SU. 2003. Expression and anti-bacterial activity of human parotid secretory protein (PSP). Biochem Soc Trans. 31:815–818.
- Gerlofs-Nijland ME, Dormans JAMA, Bloemen HJT, Leseman DLAC, Boere AJF, Kelly FJ, Mudway IS, Jimenez AA, Donaldson K, Guastadisegni C, et al. 2007. Toxicity of coarse and fine particulate matter from sites with contrasting traffic profiles. Inhalation Toxicol. 19:1055–1069.
- Gorr SU, Abdolhosseini M, Shelar A, Sotsky J. 2011. Dual host-defence functions of SPLUNC2/PSP and synthetic peptides derived from the protein. Biochem Soc Trans. 39:1028–1032.
- Guaita R, Pichiule M, Maté T, Linares C, Díaz J. 2011. Short-term impact of particulate matter (PM2.5) on respiratory mortality in Madrid. Int J Environ Health Res. 21:260–274.
- Gualtieri M, Longhin E, Mattioli M, Mantecca P, Tinaglia V, Mangano E, Proverbio MC, Bestetti G, Camatini M, Battaglia C, et al. 2012. Gene expression profiling of A549 cells exposed to Milan PM2.5. Toxicol Lett. 209:136–145.
- Guarnieri M, Balmes JR. 2014. Outdoor air pollution and asthma. Lancet. 383:1581–1592.
- Halonen JI, Lanki T, Yli-Tuomi T, Tiittanen P, Kulmala M, Pekkanen J. et al. 2009. Particulate air pollution and acute cardiorespiratory hospital admissions and mortality among the elderly. Epidemiology. 20:143–153.
- IARC. 2013. Air pollution and cancer. [accessed 2020 May 6]. http://www.iarc.fr/en/publications/books/sp161/AirPollutionandCancer161.pdf
- Ji YN, Wang Q, Suo LJ. 2012. CYP1A1 Ile462Val polymorphism contributes to lung cancer susceptibility among lung squamous carcinoma and smokers: a meta-analysis. PLOS One. 7:e43397.
- Korashy HM, El-Kadi AOS. 2005. Regulatory mechanisms modulating the expression of cytochrome P450 1A1 gene by heavy metals. Toxicol Sci. 88:39–51.
- Lee KY, Batmunkh T, Joo HS, Park K. 2018. Comparison of the physical and chemical characteristics of fine road dust at different urban sites. J Air Waste Manag Assoc. 68:812–823.
- Ministry of Environment. 2016. What is the particulate matter? [accessed 2020 May 6]. http://me.go.kr/home/file/readDownloadFile2.do?fileId=127372&fileSeq=1&fileName=968ea7e08690ebc49b8d6fc4acec5fe7a65f35aee9723b5e33bd822a90cfe874294e029bed96e3250c0b2727ef682584c50c83635c79eed6166b8be5e6af1223&openYn=Y
- Mollerup S, Berge G, Baera R, Skaug V, Hewer A, Phillips DH, Stangeland L, Haugen A. 2006. Sex differences in risk of lung cancer: expression of genes in the PAH bioactivation pathway in relation to smoking and bulky DNA adducts. Int J Cancer. 119:741–744.
- OECD. 2016. The Economic Consequences of Outdoor Air Pollution. [accessed 2020 May 6]. https://www.oecd-ilibrary.org/content/publication/9789264257474-en
- Perez L, Tobías A, Querol X, Pey J, Alastuey A, Díaz J, Sunyer J. 2012. Saharan dust, particulate matter and cause-specific mortality: A case–crossover study in Barcelona (Spain). Environ Int. 48:150–155.
- Pope III CA. 2002. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 287:1132–1141.
- Prokopovic V, Popovic M, Andjelkovic U, Marsavelski A, Raskovic B, Gavrovic-Jankulovic M, Polovic N. 2014. Isolation, biochemical characterization and anti-bacterial activity of BPIFA2 protein. Arch Oral Biol. 59:302–309.
- Przygodzki R, Bennett WP, Guinee DG Jr, Khan MA, Freedman A, Shields PG, Travis WD, Jett JR, Tazelaar H, Pairolero P, et al. 1998. p53 mutation spectrum in relation to GSTM1, CYP1A1 and CYP2E1 in surgically treated patients with non-small cell lung cancer. Pharmacogenetics. 8:503–511.
- Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, Hoffmann B, Fischer P, Nieuwenhuijsen MJ, Brunekreef B, et al. 2013. Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European Study of Cohorts for Air Pollution Effects (ESCAPE). Lancet Oncol. 14:813–822.
- Sawrycki P, Domagalski K, Cechowska M, Gąsior M, Jarkiewicz-Tretyn J, Tretyn A. 2018. Relationship between CYP1B1 polymorphisms (c.142C > G, c.355G > T, c.1294C > G) and lung cancer risk in Polish smokers. Future Oncol. 14:1569–1577.
- Sharma AK, Jensen KA, Rank J, White PA, Lundstedt S, Gagne R, Jacobsen NR, Kristiansen J, Vogel U, Wallin H, et al. 2007. Genotoxicity, inflammation and physico-chemical properties of fine particle samples from an incineration energy plant and urban air. Mutat Res. 633:95–111.
- Shimada T, Fujii-Kuriyama Y. 2004. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and1B1. Cancer Sci. 95:1–6.
- Sissung TM, Price DK, Sparreboom A, Figg WD. 2006. Pharmacogenetics and regulation of human cytochrome P450 1B1: implications in hormone-mediated tumor metabolism and a novel target for therapeutic intervention. Mol Cancer Res. 4:135–150.
- Sun H, Shamy M, Kluz T, Muñoz AB, Zhong M, Laulicht F, Alghamdi MA, Khoder MI, Chen LC, Costa M, et al. 2012. Gene expression profiling and pathway analysis of human bronchial epithelial cells exposed to airborne particulate matter collected from Saudi Arabia. Toxicol Appl Pharmacol. 265:147–157.
- Taioli E, Gaspari L, Benhamou S, Boffetta P, Brockmoller J, Butkiewicz D, Cascorbi I, Clapper ML, Dolzan V, Haugen A, et al. 2003. Polymorphisms in CYP1A1, GSTM1, GSTT1 and lung cancer below the age of 45 years. Int J Epidemiol. 32:60–63.
- Tecer LH, Alagha O, Karaca F, Tuncel G, Eldes N. 2008. Particulate matter (PM2.5, PM10-2.5, and PM10) and children’s hospital admissions for asthma and respiratory diseases: a bidirectional case-crossover study. J Toxicol Environ Health A. 71:512–520.
- Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. 2008. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 25:843–854.
- Thurston GD, Kipen H, Annesi-Maesano I, Balmes J, Brook RD, Cromar K, De Matteis S, Forastiere F, Forsberg B, Frampton MW, et al. 2017. A joint ERS/ATS policy statement: what constitutes an adverse health effect of air pollution? An analytical framework. Eur Respir J. 49:1600419.
- Uppstad H, Øvrebø S, Haugen A, Mollerup S. 2010. Importance of CYP1A1 and CYP1B1 in bioactivation of benzo[a]pyrene in human lung cell lines. Toxicol Lett. 192:221–228.
- Wapinski O, Chang HY. 2011. Long noncoding RNAs and human disease. Trends Cell Biol. 21:354–361.
- Wendling Z, Emerson J, Esty D, Levy M, de Sherbinin A. et al. 2018. 2018 Environmental Performance Index (EPI).
- WHO. 2013. Health effects of particulate matter. Policy implications for countries in Eastern Europe, Caucasus and central Asia. [accessed 2020 May 6]. http://www.euro.who.int/__data/assets/pdf_file/0006/189051/Health-effects-of-particulate-matter-final-Eng.pdf?ua=1
- Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. 2015. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 28:529–540.
