Abstract
This was a pilot study carried out to develop a new protein food item from imbibed soybean before germination. It identified the significance of a short stage after imbibition and before germination, and that vitamin C production was activated in as little as 16 h from the start of imbibition, without any influence on the soy protein quality or sensory acceptability, while longer imbibition caused the imbibed soybean to activate its phytophysiological metabolism for germination. DNA microarray analysis indicated that the genes for carbohydrate metabolism were up-regulated prior to 16 h, and that the expression rates of genes responsible for environmental factors were down-regulated. Thereafter, the expression rates of the genes associated with lipid metabolism and secondary metabolite production were changed. This information should contribute to a better understanding of how to develop a new soy protein item in pre-germination before active physiological processes begin.
Soybean as a highly nutritious food material contains a variety of functional components, including isoflavones, oligosaccharides, and dietary fiber.Citation1) It has been consumed as a vegetable protein in Japan from long ago. It is indispensable for making soybean curd (tofu), fermented soybean paste (miso), and soy milk after soaking of beans in water overnight prior to germination.
Germination is regulated by genes as well as by environmental factors.Citation2,Citation3) In phase I, imbibition is initiated by physical absorption of ambient moisture. Metabolism is reactivated by enzymes that were stored in the seed during maturation. In phase II, proteins are synthesized by new mRNAs, with no increase in water absorption. Development of the seedling takes place at phase III, in which the embryonic axes elongate and stored reserves are mobilized in the beans.Citation2) The genes and substances involved in germination have been identified for Arabidopsis and Medicago by many molecular and physiological studies.Citation4,Citation5) Recently, the total genome sequences of the soybean were elucidated and published as databases.Citation6) Transcriptomics and proteomics of the soybean have revealed the metabolic changes in the sprouts. Nanjo et al.Citation7) found that 6 or 12 h of flooding stress for 2-d-old seedling soybeans affected the expression of numerous genes related to photosynthesis, glycolysis, amino acid metabolism, protein metabolism, secondary metabolism, etc. in the roots, including the hypocotyls. On the other hand, transcriptome analysis of cotyledons from the yellow (storage) to the green (photosynthetic) stage, at which the plant is 7–8 cm long, revealed that changes in expression of elements of the glyoxylate pathway play a fundamental role during the functional transition of the cotyledon from nutrient storage to photosynthesis.Citation8) No information is available regarding the process after imbibition of 1 d or a few hours (phase I–III), after which transcriptomes and metabolomes begin to change. Analysis of the proteome profile at 12 h after soybean imbibition indicated that lipids and proteins were degraded through the lipoxygenase-dependent pathway and the protease and 26S proteosome systems, respectively.Citation9) Little is known of time-course changes in food functionality-related transcriptomics during very short-term imbibition.
In the field of food science, it has long been known that 12–72 h of germination of soybean, rice, and broccoli produces vitamin C, γ-aminobutyric acid (GABA), and isothiocyanate;Citation10−Citation12) but, before and while these components increase, research has not focused on what genes are expressed or what metabolism is altered around component changes. The importance of studying short-term imbibition was, thus, emphasized to produce a new soy protein material. In the present study, we paid special notice to imbibition as little as 0–16 h, preceding seedling growth and differentiation to the soybean plant. We conducted DNA microarray analysis to examine changes in the expression of genes coding for factors that may determine food protein quality.
Material and methods
Experimental design
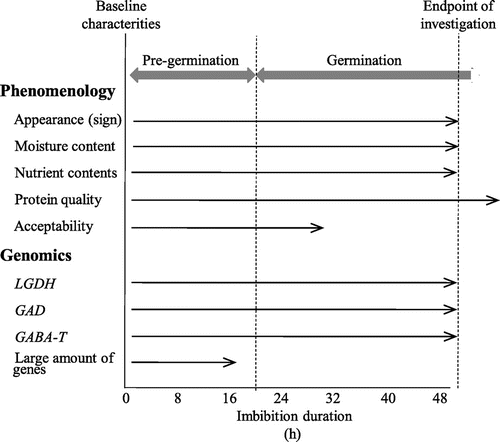
As for the phenomenology of short-term imbibition, the moisture content, nutrient content, protein quality, and sensory acceptability were examined (Fig. ). Genomics research of imbibed soybeans was conducted to support the phenomenological data. Also, time-course changes in genes related to vitamin C and GABA metabolism were compared by quantitative real-time polymerase chain reaction (PCR) analysis.
Soybean seeds
Thirty dry seeds of Glycine max (cv. Suzumaru) were soaked in 1% hypochlorous acid and then washed in water. They were placed on moistened absorbent cotton and allowed to germinate in the dark at 25 °C. The condition of 8-h-imbibed seeds was beans imbibed more than 0.06 g of water. After 16 h of imbibition, seeds that were germinated or imbibed more than 0.33 g were used for sample. The 24-, 32-, 40-, and 48-h-imbibed seeds were collected, and they grew to a radicle length of more than 2, 5, 15, and 20 mm, respectively. About 20–25 seeds at each imbibition stage were collected, trichotomized, and milled with dry ice individually. All the samples were stored at −80 °C prior to use.
Vitamin C and GABA determination
Vitamin C was measured by a colorimetric method using 2,4-dinitrophenylhydrazine. In brief, 0.5 g of the frozen sample was homogenized in 6 mL of 5% metaphosphate and this was centrifuged at 5000 rpm for 10 min at 4 °C. Next, 0.2% 2,6-dichlorophenol-indophenol sodium salt dihydrate was added to 2 mL of the supernatant, the maximum amount at which the pink color did not disappear. Two mL of 1% tin(II) chloride and 1 mL of 2% 2,4-dinitrophenylhydrazine/sulfuric acid (N/10) were added to the solution, and this was incubated for 3 h at 37 °C. Then, 5 mL of 85% sulfuric acid was added gradually. The solution was incubated for 30 min at room temperature and the absorbance of 540 nm was measured. For the blank, 1 mL of 2% 2,4-dinitrophenylhydrazine/sulfuric acid (N/10) was added to that supernatant 30 min prior to measurement of the absorbance.
To determine the GABA content of each sample, a frozen sample was homogenized with water and centrifuged at 15,000 rpm for 3 min at 4 °C. The supernatant was dried under reduced pressure. Then, 0.3 mL of 0.1 M Na2CO3-NaHCO3 buffer (pH 8.9) and 0.3 mL of 10 mM dansyl chloride-acetone were added and mixed in well, and this was incubated at 70 °C for 6 min. The reacted solution was centrifuged at 10,000 rpm for 10 min at 4 °C, and the supernatant was analyzed for GABA content by an HPLC equipped with a spectrophotometric detector at 436 nm. A CrestPak C18S column (Jasco Corp., Tokyo, 150 × 4.6 mm) was used. The column temperature was maintained at 25 °C, and the flow rate was set at 1.4 mL/min. The column was equilibrated with 80% solvent A (10 mM citrate buffer solution pH 6.5–4% N,N-dimethylformamide) and 20% solvent B (30% solvent A-70% acetonitrile), and then elution was done with a 20–70% solvent B gradient.
Protein quality as glucono-δ-lactone-induced coagulation and sensory evaluation of soymilk
Zero-, 16-, 32-, and 48-h-imbibed soybeans (60 g each) were added to 300 mL of water, homogenized for 1 min, and then heated for 8 min at 100 °C to stop some endogenious enzyme activities. The solution was filtered with gauze and filled to 240 mL with water.
Degrees of degradation of seed storage protein due to imbibition were estimated by adding glucono-δ-lactone to soymilk, which was diluted with water 50-fold to a final concentration of 3.5%.
Sensory evaluation of the soymilk was conducted by 72 panelists of Tokyo University of Agriculture. Each panelist evaluated 4 test samples (0, 16, 36, and 48 h) in random order and rinsed the mouth with water freely. Eleven-point scales from 0 (extremely distasteful) to 10 (extremely preferable) were used. Each sample (10 mL) in a cup was labeled by 3-digit random number and was kept at 10 °C prior to being served. For statistical data analysis, outliers were removed and the remaining data were used. The Tukey–Kramer HSD multiple comparison test was done for the sensory evaluation data. Data analysis was carried out with statistical software JM9.0.3 (SAS Institute, Cary, NC).
Quantitative real-time PCR analysis
Total RNA (3 samples/stage) was extracted from the frozen samples with an RNeasy plant mini kit (Qiagen, Tokyo) and purified using the same kit. RNA quality and quantification were checked with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA) and NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). One μg of total RNA from each sample was used as template for first-strand cDNA synthesis with oligo(dT) primer with a first-strand cDNA synthesis kit (GE Healthcare, Tokyo). Quantitative real-time PCR was done on a thermal cycler dice (Takara, Tokyo) using SYBRPremix Ex Taq (Takara, Tokyo) following the manufacturer’s protocol. Primer sets were designed with a Primer3 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) as follows : l-galactono-1,4-lactone dehydrogenase (LGDH), 5′-CGCATATGAACACTGGGCTA-3′ and 5′-ATTGGGGTCCAATTCCTTTC-3′, GenBank accession No. XM_003535854; glutamate decarboxylase (GAD), 5′-GACTTCTTAAGGCGCTTTGG-3′ and 5′-GCAGAACCTTCTCAACATCG-3′, NM_001254052; GABA transaminase (GABA-T), 5′-TCCTGAATGGGGAATAGGTG-3′ and 5′-CATGATGTATCTCCCGCAAC-3′, AK244308; actin, 5′- TGAGCGTTTCAGATGTCCTG -3′ and 5′- GGTGCTGAGAGAAGCCAAGA -3′, AK285936. PCR was carried out as follows: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The amounts of GAD, GABA-T, and LGDH expression were normalized to the level of actin expression. Relative amounts of gene expression were calculated against the amount of dry seeds by the standard curve method.
DNA microarray analysis
Total RNAs (200 ng) of 0-, 8-, and 16-h-imbibed samples were amplified with a low RNA Input Quick Amp Labeling Kit fluorescent linear amplification kit, one-color (Agilent Technologies, Palo Alto, CA). Cy3-labeled cRNA (1.65 μg) was fragmented to an average size of about 50–100 nucleotides and then hybridized for 17 h at 65 °C to a soybean oligonucleotide microarray slide, which was washed and scanned in the Cy3 channel with an Agilent DNA microarray scanner (Agilent Technologies, USA). Signal intensity per spot was generated from the scanned image with Agilent scan control 8.0 software by Agilent Technologies’ standard operation procedures. Spots that did not pass quality control procedures were flagged as “Not Detected.” The scanned images were analyzed with a commercial software (Gene spring GX11.5) as follows: (i) If gene expressions for more than 7 of 8 samples were “Not detect,” the gene was excluded, (ii) threshold raw signals were set at unify, and (iii) submitted to 75th percentile normalization. Hierarchical clustering was then done by pvclust functionCitation13) in statistical language R,Citation14) version 2.14.0. Differences were found to be significant by one-way ANOVA (p = 0.01), followed by a Tukey’s HSD post hoc test. Fold change greater than 5-fold was used to compare the expression patterns of genes adjacent stages. The experiment was duplicated. All microarray data were submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, Series ID GSE48913; http://www.ncbi.nlm.nih.gov/geo/).
Gene ontology analysis
Two tips for 0 h sample and 3 tips of the 8 h and 16 h samples were used for the following analysis. To obtain gene ontology (GO) terms on the identified genes, the Arabidopsis Information Resource (TAIR) locus ID was assigned to each soybean probe using Biomart (http://plants.ensembl.org/biomart/). Then, TAIR IDs were uploaded to the DAVID website (http://david.abcc.ncifcrf.gov/). The functional annotation chart was analyzed on the basis of biological processes in GO, GOTERM_BP_ALL. To extract the statistically over-represented GO terms in each group of differentially expressed genes and to correct the results by multiple testing, we used EASE scores, which modified Fisher’s exact test p values,Citation15) and Benjamini and Hochberg false discovery rate (FDR) corrections.Citation16) The Benjamini and Hochberg FDR-corrected p value of < 0.05 indicated a significantly enriched GO term. To determine the hierarchical structure of the selected GO terms, we used QuickGO (https://www.ebi.ac.uk/QuickGO/), an online analysis utility.
Results
Imbibition-induced phenomenological changes and the formation of vitamin C and GABA
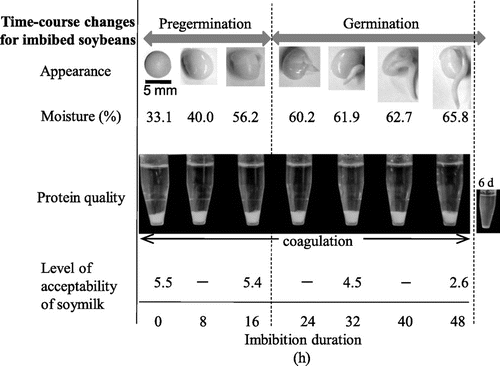
After imbibition, the moisture of the seeds increased for up to 24 h after imbibition, and stabilized thereafter (Fig. ). At the end of moisture increase, germination was complete.
Fig. 2. Time-course changes of imbibed soybeans during pre-germination and germination.
Notes: Protein quality is represented as glucono-δ-lactone-induced protein precipitability. Acceptability was measured with soymilk prepared from imbibed beans.
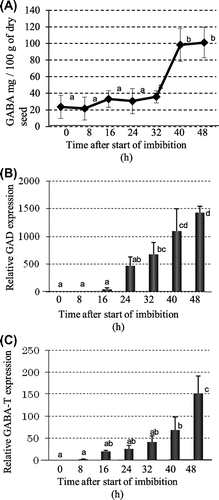
The vitamin C content of dry soybean seeds was 1.6 mg/100 g and increased 2-fold after 8 h of imbibition (Fig. (A)). The concentration of vitamin C increased significantly within 16 h of imbibition, and the increase was constant through imbibition. By real-time PCR analysis, the expression level of LGDH, which catalyzes the last step of the putative vitamin C biosynthetic pathway, was the same between dry seeds and 8 h of imbibition, although vitamin C increased 2-fold within 8 h of imbibition (Fig. ). The relative quality of the LGDH genes largely increased during 48 h of imbibition.
Fig. 3. Time-course changes in Vitamin C and related genes after imbibition.
Notes: (A) Concentration of vitamin C; (B) Gene expression of ascorbate synthesizing enzyme l-galactono-1,4-lactone dehydrogenase (LGDH).
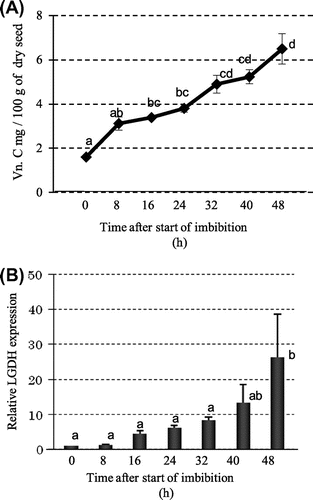
The GABA content of the dry soybean seed was 23.6 mg/100 g seeds and increased slightly until 32 h of imbibition (Fig. (A)). A large increase in GABA in the beans occurred 32–40 h after imbibition, from 35.8 to 98.3 mg/100 g of seeds. GABA is synthesized mainly by GAD from glutamate and is degraded by GABA transaminase. Soybean has one copy of the GAD gene,Citation17) and real-time PCR showed a 2.7-fold increase within 8 h of imbibition and a 37.2-fold increase within 16 h (Fig. (B)). GABA transaminase also increased 2.3-fold by 8 h of imbibition, and further increased thereafter (Fig. (C)).
Protein quality and acceptability of soymilk using imbibed soybeans
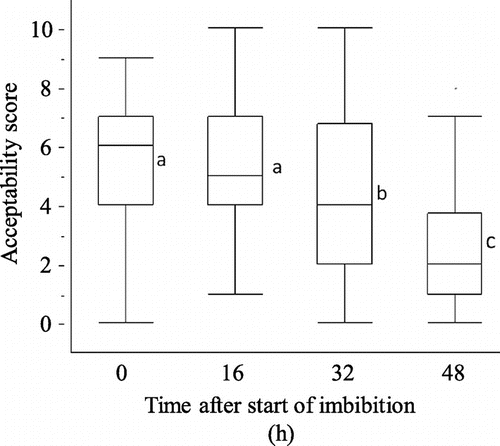
Glucono-δ-lactone-induced protein precipitability showed that the precipitability of the protein was not changed by 48 h of imbibition (Fig. ). Coagulation did not appear in the 6-d-imbibed soy protein. Acceptability were similar between the dry beans and the 16-h-imbibied beans, while the samples imbibed for 32 h and longer showed significant unacceptable taste at p < 0.05 (Fig. ). The acceptability of soymilk is worse after drastic gene expression changes, although the concentrations of GABA increased 32 h later. Here, we carried out a DNA microarray analysis with 0-, 8-, 16-h-imbibed seeds.
Numbers of differentially expressed genes at the 3 different stages of imbibition
A total of 42,780 genes set in a chip were investigated. We normalized 3 stages and 8 samples by 75th percentile shift, and constructed a cluster dendrogram (Fig. (A)) with the result that the original dry seed and the 8-h-imbibed seed belonged to the same cluster, whereas the 16-h-imbibed sample was found to be belonging to a different cluster. After statistical analysis as described above in “Materials and Methods,” 7792 genes were extracted.
Fig. 6. Cluster dendrogram for genes expressed in the soybean.
Notes: (A) Eight samples from 3 imbibition stages constructed using the pvclust function; (B) Number of differentially expressed genes extracted at fold change > 5 as compared between 2 adjacent stages. The number of all differentially expressed genes, overlapping eliminated, was 1160.
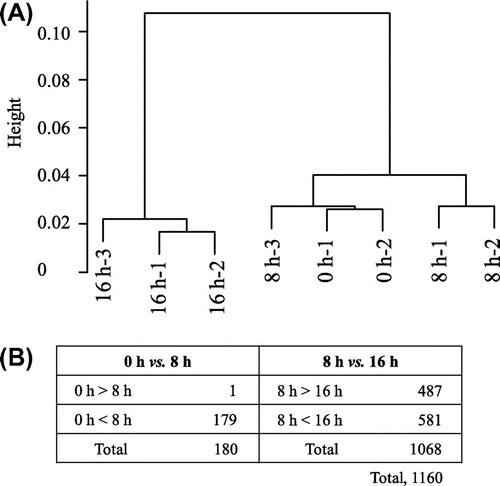
The between-stage difference of more than 5-fold was used to extract 1160 genes (Fig. (B)). The genes with such a fold difference between the 0-h- and 8-h-imbibed seeds amounted to 180. These were up-regulated genes except for 1 gene and between the 8-h- and 16-h-imbibed seeds amounted to 1068, which included 487 and 581 down- and up-regulated genes, respectively.
Classification of different expressed genes by expression patterns
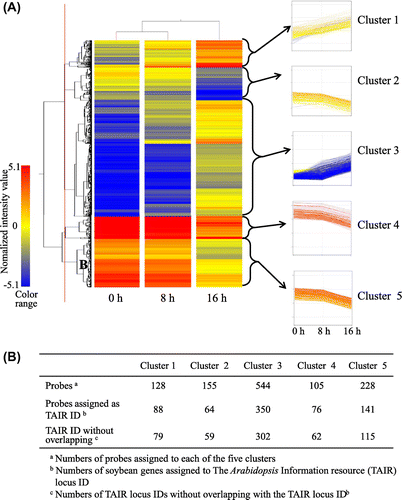
The 1160 genes were classified into the 5 expression patterns (Fig. ): cluster 1128 genes that increased in expression during imbibition; cluster 3, 544 genes that increased during 0–8 h or 8–16 h after imbibition; cluster of 2, 4, and 5, which contained 155, 105, and 228 genes, showing drastic decreases in expression between 8 and 16 h after imbibition, though the expression levels of genes in the dry seeds were different. Detailed microarray data are shown in the supplemental table.
GO enrichment analysis
By GO analysis of the above 5 clusters, we found significantly enriched the GO terms (Table ). The GO term of the categories appearing deepest in the hierarchy is more specific and an enrichment GO terms.
Table 1. Significantly enriched GO terms found in the groups.
Cluster 1 made major contributions to response to metal ions, such as phosphoenolpyruvate carboxykinase and copper chaperone, which need Mg2+ or K+ as a cofactor. Cluster 1 was also enriched related to oxidation-reduction process, such as cytochrome P450, ascorbate peroxidase, and malate dehydrogenase. Cluster 3 genes were enriched for the cellular carbohydrate metabolic processes, such as omega-3 fatty acid desaturase and xyloglucan endotransglycosylasehydrolase. Cell wall organization was also enriched GO term, which contained cellulose, pectin methylesterase, xyloglucan fucosyltransferase, and lipid transfer protein. Cellular amino acid derivative biosynthetic process is also significantly enriched; it is related to alkaloid and flavonoid biosynthesis enzymes, not among the top 3 significantly enriched GO terms. The genes related to jasmonic acid biosynthetic process were also enriched. These encode lipoxygenase (LOX). In clusters 2, 4, and 5, there were genes with significant expressions in response to water deprivation, which contained LOX2, response to temperature stimulus, response to light intensity, and response to abscisic acid stimulus.
Gene expression profile with respect to food-related genes
Almost all the genes encoding proteases were up-regulated (Table ). Glycinin rapidly decreased after 72 h of imbibition, whereas basic chains of glycinin and β-conglycinine were detectable at least after 120 h of growth,Citation18) though no information is available regarding changes in proteins and amino acids during short-term imbibition of soybean. In our experiment, we found that almost all the genes for proteases, including metallo, serine, and cysteine proteases, existed until 8 h after imbibition and up-regulated at 16 h of imbibition, whereas aspartic proteases and some of the serine and cysteine proteases were down-regulated at 8–16 h of imbibition. Genes for Bowman-Birk and cystein protease inhibitors were down-regulated at 8–16 h of imbibition. The Kunitz protease inhibitors were up-regulated at this imbibition stage.
The gene for glucose producing cellulose existed after 8 h of imbibition, and was up-regulated thereafter. The genes of β-glucosidase also produced glucose, and were regulated during 8–16 h imbibition. The genes of sucrose synthase were up-regulated 8–16 after imbibition.
Soybean LOX1, 2, and 3 have been studied extensively because their activity in the seed results in an unpleasant flavor.Citation19) α-Linoleic acid is degraded by reaction with LOX and hydroperoxide lyase with the formation of hexanal.Citation20) After 16 h of imbibition, GLYMA13G42320 (LOX1), GLYMA13G42310 (LOX2), GLYMA07G00920, and GLYMA15G03030 (LOX3) were down-regulated and the gene for hydroperoxide lyase was kept almost constant. Oxidation of polyunsaturated fatty acids by LOXs might be involved in the β-oxidation step of jasmonic acid biosynthesis, whose pathway was up-regulated by GO analysis (cluster 3).
Other anti-nutritional genes, squalene monooxygenase, involved in saponin synthesis appeared in the 16-h-imbibed seed, while the amount of saponin doubled 1 d after imbibition and then showed no change until 2 d.Citation21) Lectin gene was up-regulated at 8–16 h of imbibition. Phytic acid is synthesized from glucose-6-phosphate, which is converted to myo-inositol 3-phosphate by myo-inositol-1-phosphate synthase (MIPS). MIPS was down-regulated at 8–16 h of imbibitions, whereas the phytic acid content in the seeds increased by 24 h of imbibition.Citation22) Imbibition of soybean induces the production of isoflavons by synthase and dehydrogenase, with a change to phytoallexin by isoflavone reductase. Isoflavon synthase expression did not change, whereas the isoflavon reductase gene underwent a large degree of up-regulation. The gene encoding ferritin was up-regulated at 8–16 h of imbibition.
Discussion
The gene expression profile of short-term imbibed soybeans before germination and the germinating processes was examined by DNA microarray analysis. Transcript analysis characterized the gene expression profiles of 16-h-imbibed soybean and GO analysis revealed that there were genes for carbohydrate metabolism causing changes in nutrients, taste, and food functions which were up-regulated after 8 h of imbibition (cluster 3). On the other hand, genes related to phytophysiological processes such as carbon fixation and oxidoreduction were up-regulated during imbibition (cluster 1). Genes expressed during embryogenesis and translated to proteins for break dormancy were degraded after 8 h of imbibition (clusters 2, 4, and 5).
Peptide and proteinogenic amino acids have sweet, bitter, sulfurous, and umami tastes. Focusing on genes that may determine the food-processing properties, proteases and inhibitors-coding genes had been up- or down-regulated during 16 h of imbibition. Mizuno et al.Citation23) have reported that the amount of free amino acids at baseline is similar to that at the 24-h-imbibition stage and that the amount doubles at the 48-h-imbibitions stage. Thus, genes for proteases were regulated during short-term imbibition before changes in the amounts of free amino acids in soybean. Tan-Wilson et al.Citation24) observed that Kunitz-type protease inhibitor expressed 1 week after the start of imbibition followed by its gradual down-regulation. Papastoitsis et al.Citation25) indicated that a 29,000 Da cysteine protease acted to degrade Kunitz and Bowman-Birk trypsin inhibitors. These data suggest that the genes related to the synthesis and degradation of proteins became active during 8–16 h of imbibition in soybeans. Glycinine and β-conglycinin are major seed proteins in soybean, and glycinin rapidly decreased after 72 h of imbibition, whereas no obvious decrease in β-conglycinin was detectable at 48 h of imbibition.Citation18) Glucono-δ-lactone-induced protein precipitability did not change until 6 d after imbibition. Imbibition for up to 48 h did not change molecular weight, as SDS-PAGE of soybean showed (Supplemental Fig. 1; see Biosci. Biotechnol. Biochem. website), whereas application of Molsin, an Aspergillus saitoi protease in soybean curd, changed the sizes of the bands to 20 kDa or less,Citation26) and the curd did not show any indisputable rupture point, having a cream cheese-like texture. This suggests that the processability of soybean is not affected by short-term imbibition. At the imbibition stage between phase I and phase II, 8–16 h of imbibition, components related to sensory acceptability of soybean underwent little change, although many genes including cellulose, β-glucosidase and sucrose synthase, were regulated during 8–16 h imbibition (Fig. , Table ). Expression changes in anti-nutritional genes, LOX, squalene monooxygenase, lectin, etc. should cause significant unacceptable taste of soy milk made from 32-h-imbibed seeds.
Table 2. Gene titles related to food function and expression changes compared between 2 adjacent stages.
The vitamin C content of the soybean seeds increased 2-fold after 8 h of imbibition, whereas the LGDH gene significantly increased after 48-h imbibition. These results suggest that the vitamin C increase within 8 h of imbibition was due to the activity of the LGDH enzyme, which existed in the dry seeds (Supplemental Fig. 1). Vitamin C-degrading enzymes have not been identified in soybean, but 3 ascorbate oxidases and 8 ascorbic acid peroxidases have been found in Arabidopsis (TAIR, http://www.arabidopsis.org/). In our microarray analysis of soybeans, genes with similar sequences were up-regulated 8–16 h after the start of imbibition. The other genes for enzymes related to vitamin C metabolism were not greatly changed during 16-h imbibition.
GABA is formed by GAD from glutamic acid, GAD is activated by increases in the cytosolic concentration of H+ or Ca2+, and the cytosolic levels of Ca2+ are elevated in response to osmotic stress.Citation27) The GABA contents of soybean and rice were increased by flood.Citation7,Citation17) GABA content of soybeans increased slightly until 32 h of imbibitions, by which gene encoding GAD increased and GAD is activated after 32 of imbibition. PCR for GABA biosynthetic and metabolic enzymes was carried out using the primers shown in legend to supplemental Fig. 2; see Biosci. Biotechnol. Biochem website; GABA-T was detected in the dry seeds, whereas no GAD-specific fragment was detected in them during 0–8 h of imbibition. Hence, it was suggested that the relative changes in mRNA expression of GAD after imbibition were larger than that of GABA-T. In our microarray analysis, the genes for glutamate synthetase, which produce glutamate and ammonia from glutamine, were also up-regulated at 8–16 h of imbibition. GABA is also synthesized by oxidation of polyamine, whose functions are related to cell cycle, cell division, growth, as well as to stress phenomena.Citation28) One of the triamines, spermidine, was oxidized by polyamine oxidase and diamine oxidase and converted to GABA. Diamine oxidase was expressed in the beans at 16 h of imbibition.
These data indicate that the global transcriptomics of the soybean during imbibtion is active and dynamic. We can see high gene expression of chemical stimulus-related and cellular carbohydrate metabolic process-related genes involved in soybean imbibition. The transcriptomics also delineate the expression changes in non-nutrient and taste-related genes under short-term imbibition. A soy protein product with increased functionality can thus be produced in as little as 16-h imbibition of soybean. This information might open the way to future studies for a better understanding of how to develop a new soy protein item in pre-germination before active phytophysiological processes begin.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.877822.
Supplemental Table - Identification of 1160 Putative Soybean Reference Genes Based on Agilent Analysis of Short Term Imbibition
Download MS Excel (453 KB)Supplemental Figs. 1-2
Download PDF (270.3 KB)Acknowledgment
This study was supported by a research grant from the Fuji Foundation for Protein Research.
Notes
Abbreviations: FDR, false discovery rate; GABA, γ-aminobutyric acid; GABA-T, GABA transaminase; GAD, glutamate decarboxylase; GEO, gene expression omnibus; GO, gene ontology; LGDH, l-galactono-1,4-lactone dehydrogenase; LOX, lipoxygenase; MIPS, myo-inositol-1-phosphate synthase; NCBI, National Center for Biotechnology Information; PCR, polymerase chain reaction; TAIR, The Arabidopsis Information Resource.
References
- Anderson RL, Wolf WJ. J. Nutr. 1995;125:581–588.
- Bewleyl JD. Plant Cell. 1997;9:1055–1066.
- Weitbrecht K, Müller K, Leubner-Metzger G. J. Exp. Bot. 2011;62:3289–3309.
- Rajjou L, Gallardo K, Debeaujon I, Vanderkerckhove J, Job C, Job D. Plant Physiol. 2004;134:1598–1613.
- Buitink J, Leger JL, Guisle I, Vu BL, Wuilleme S, et al. Plant J. 2006;47:735–750.
- Schmutz, J, Cannon SB, Schlueter J, Ma J, Mitros T, et al. Nature. 2010;463:178–183.
- Nanjo Y, Maruyama K, Yasue H, Yamaguchi SK, Shinozaki K, et al. Plant Mol. Biol. 2011;77:129–144.
- Gonzalez DO, Vodkin LO. BMC Genomics. 2007;8:468.
- Han C, Yin X, He D, Yang P. PLOS ONE. 2013;8:e56947.
- Wai KNT, Bishop JC, Mack PB, Cotton RH. Plant Physiol. 1947;22:117–126.
- Komatsuzaki N, Tsukahara K, Toyoshima H, Suzuki T, Shimizu N, et al. J. Food Eng. 2007;78:556–560.
- Cartea ME, Velasco P. Phytochem. Rev. 2008;7:213–229.
- Suzuki R, Shimodaira H. Bioinformatics. 2006;22:1540–1542.
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2006.
- Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA. Genome Biol. 2003;4:R70.
- Benjamini Y, Hochberg Y. J. R. Stat. Soc. Ser. B. 1995;57:289–300.
- Matsuyama A, Yoshimura K, Shimizu C, Murano Y, Takeuchi H, et al. J. Biosci. Bioeng. 2009;107:538–543.
- Kim HT, Choi UK, Ryu HS, Lee SJ, Kwon OS. Biochim. Biophys. Acta 2011;1814:1178–1187.
- Kitamura K, Kumagai T, Kikuchi A. Jpn. J. Breed. 1985;35:413–420.
- Matoba T, Hidaka H, Narita H, Kitamura K, Kaizuma N, et al. J. Agric. Food Chem. 1985;33:852–855.
- Jyothi TC, Kanya TCS, Rao AGA. J. Food Biochem. 2007;31:1–13.
- Egli I, Davidsson L, Juillerat MA, Barclay D, Hurrell RF. J. Food Sci. 2002;67:3484–3488.
- Mizuno T, Yamada K. Nippon Shokuseikatugaku Kaishi. 2007;17:51–57. Japanese.
- Tan-Wilson AL, Rightmire BR, Wilson KA. Plant Physiol. 1982;70:493–497.
- Papastoitsis G, Wilson KA. Plant Physiol. 1991;96:1086–1092.
- Nishinoaki M, Asakura T, Watanabe T, Kunizaki E, Matsumoto M, et al. Biosci. Biotechnol. Biochem. 2008;72:587–590.
- Kinnersley AM, Turano FJ. Crit. Rev. Plant Sci. 2000;19:479–509.
- Edreva A. Bulg. J. Plant Physiol. 1996;22:73–101.