Abstract
The unicellular red alga Cyanidioschyzon merolae is used as a model organism to investigate the basic architecture of photosynthetic eukaryotes. We established a stable expression system for the green fluorescent protein fused with the phycocyanin-associated rod linker (APCC) protein in C. merolae, which was clearly localized on the plastid. This system should be useful in the genetic engineering of C. merolae.
Cyanidioschyzon merolae is a unicellular red alga found in acidic hot springs. The cell contains one nucleus, one mitochondrion, and one plastid, each of which has its own genome.Citation1) The complete sequences of these three genomes have been determined.Citation2–5) It has been proposed that C. merolae is a primitive phototrophic eukaryote in view of its extremely simple cell structure and minimally redundant small genome. Hence, it is used as a model organism to investigate the basic architecture of photosynthetic eukaryotes. In addition, there is increasing interest in C. merolae because it converts solar energy to biomass even under hot acidic conditions, and thus it might be useful in the production of biofuels, reducing the risk of environmental contamination. We have reported several genetic engineering methods for C. merolae, including transient gene expression,Citation6) a green fluorescent protein (GFP) reporter system,Citation7) transient gene suppression,Citation8) and gene knockout.Citation9) These methods were utilized to determine the novel mechanisms utilized by C. merolae. In this study, we newly established a stable expression system for exogenous genes in C. merolae.
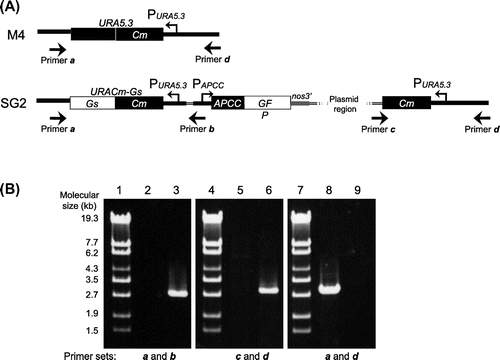
In order to establish a stable expression system for C. merolae, first we constructed integration plasmid pKF-CG2 to express C-terminally GFP-tagged APCC protein (APCC-GFP) (Fig. (A)). The APCC (ORF ID: CMO250C) gene encodes the phycocyanin-associated rod linker, a plastid light-harvesting phycobilisome protein. It is highly expressed under the light growth conditions, and the APCC-GFP protein is localized in the plastid.Citation7) We introduced pKF-CG2 into the C. merolae M4 strain, which harbors a frame-shift mutation in the URA5.3 gene (CMK046C) and thus shows uracil-dependent growth.Citation10) After polyethylene glycol-mediated transformation,Citation6) the culture was incubated for 3 d in modified Allen’s 2 (MA2) liquid medium with 5% CO2 bubbling. The culture was diluted from 10- to 104-fold, spotted on MA2 plates with embedded cornstarch,Citation11,12) and incubated for one month in a CO2 incubator at 43 °C with 5% CO2 and continuous illumination (50 μmol photon m−2 s−1). After the appearance of transformant colonies on the MA2 plates (Fig. (B)), we chose several single colonies and cultivated them in liquid MA2 medium with 5% CO2 bubbling for 10 d. Screening of the cultures for GFP fluorescence revealed that 50% of the C. merolae cells did not fluoresce (Supplemental Fig. 1(A)–(C); see doi:10.1080/09168451.2014.877823), indicating that non-transformed cells were also in the culture. Hence, the single-colony isolation step was repeated thrice. Although non-transformed cells were observed in the second isolation, we finally obtained a clone that showed GFP fluorescence in all cells (Supplemental Fig. 1(D)–(F)). In this strain, named SG2, GFP fluorescence was clearly observed in the plastid (Fig. (A)), as well as in the transformants of pCG2.Citation7) Immunoblot analysis of the SG2 strain with an anti-GFP antibody showed the expression of APCC-GFP (Fig. (B), right), but not in the wild-type strain (Fig. (B), left). Since pKF-CG2 contained a 1.4-kb fragment homologous to the sequence around the URA5.3 gene in the M4 genome, we predicted that the plasmid was integrated into this region. In order to investigate the locus in which pKF-CG2 was integrated, we extracted genomic DNA from the SG2 strain and confirmed its location by PCR. PCR products were observed when we used primers that annealed to pKF-CG2 and the genome around the URA5.3 gene (Fig. (A), primer sets a and b, c and d) in the SG2 strain (Fig. (B), lanes 3 and 6), but not in the M4 strain (Fig. (B), lanes 2 and 5). In contrast, PCR products were observed with primers annealing to the up- and downstream regions of the URA5.3 gene in the M4 strain genome (primers a and d) (Fig. (B), lane 8), but not in the SG2 strain (Fig. (B), lane 9). Perhaps the expected fragment (7.2 kb) was too long to be amplified by PCR. These results suggest that pKF-CG2 was integrated into the URA5.3 locus in the SG2 genome. Although the SG2 strain was maintained in liquid MA2 medium for more than two years at 43 °C under continuous light illumination with shaking, GFP fluorescence was still observed in all cells. In addition, this strain can be maintained by cryopreservation storage,Citation11) suggesting that this construct is highly stable in C. merolae cells.
Fig. 1. Transformation of C. merolae Cells.
Note: (A) Structure of the pKF-CG2 plasmid, which carries the URACm-Gs and APCC-GFP genes under their own promoters. It was constructed as follows: pCG2 plasmid DNA, which carries the APCC promoter (Papcc), the APCC (ORF ID: CMO250C)-GFP gene (Genbank ID: 332144728), and the polyadenylation signal sequence from the nopaline synthase gene (nos3′),Citation7,12) was digested with HindIII and treated with T4 DNA polymerase to make it blunt-ended. After digestion with EcoRI, the DNA fragment including Papcc, APCC-GFP, and nos3′ was purified. The plasmid vector DNA of pKFURACm-GsCitation13) was digested with SacI and treated with T4 DNA polymerase. After digestion with EcoRI, it was ligated with the APCC-GFP fragment. (B) Single-colony isolation from the transformant culture. The transformant culture (M4TF), made by introducing pKF-CG2 into the M4 strain, was diluted to the appropriate density and spotted on MA2 plates with embedded corn starch, and incubated for one month.
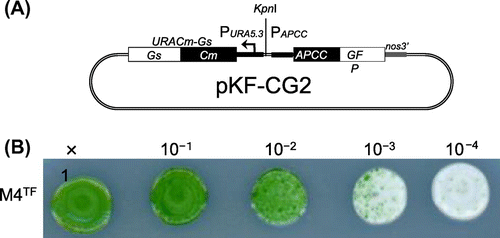
Fig. 2. Expression of the APCC-GFP Protein in C. merolae Cells.
Note: (A) Subcellular localization of APCC-GFP in the SG2 strain. Non-dividing (top) and dividing cells (bottom) were examined by fluorescence microscopy. From left to right, the images show phase-contrast microscopy, intrinsic chlorophyll fluorescence, fluorescence of GFP proteins, and superposition of chlorophyll and GFP fluorescence. Scale bar, 1 μm. (B) Immunoblot analysis with antiserum against GFP. Crude extracts were prepared from the C. merolae wild-type and SG2 cultures. Samples (20 μg for each lane), prepared as described previously,Citation6) were analyzed with mouse antiserum against GFP (MBL International, Boston, MA) as primary antibody and a horse radish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (GE Healthcare Bio-Sciences) as secondary antibody.
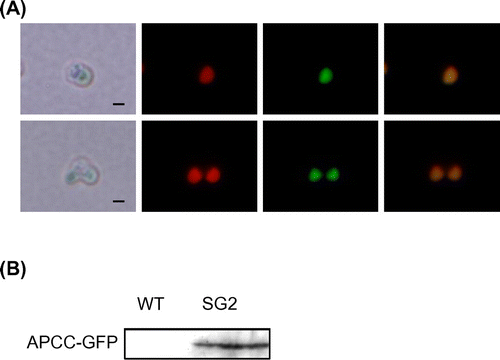
Fig. 3. Genomic Structure of the SG2 Strain.
Note: (A) Genomic structure around the URA5.3 gene in the M4 (top) and SG2 (bottom) strains. The annealing points of the primers are shown by arrows. (B) Agarose gel electrophoresis of the PCR products derived from the URA5.3 locus in the M4 (lanes 2, 5, and 8) and SG2 strains (lanes 3, 6, and 9). PCR was carried out with primer a (5-GCCGACGCTTGCTTCTGCCCATTAGG-3) and primer b (5-TCAAAAGACGGTGGTGCAGCGAGCGC-3) (lanes 2 and 3), primer c (5-ACGACGTTGTAAAACGACGGCCAGT-3) and primer d (5-ATGCACGGTGTCCAATAGGAGGAGGTCTCC-3) (lanes 5 and 6), and primers a and d (lanes 8 and 9) by ExTaq DNA polymerase (TaKaRa, Shiga, Japan). Lanes 1, 4, and 7, molecular size markers (λ-EcoT14 I digest).
In this study, we developed a gene expression system for C. merolae. We found that the M4 strain, the pKFURACm-Gs plasmid, and the APCC promoter are available for a stable gene expression system in C. merolae. C. merolae cells can integrate pKFURACm-Gs into their genome by homologous recombination around the URA5.3 gene. This stable gene expression system can be used not only for functional analysis of essential genes but also as a genetic engineering tool in C. merolae to produce valuable biomass. It has extensive possibilities for the future.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.877823.
Supplemental Figure 1. Expression of the APCC-GFP protein in C. merolae culture before (A-C) and after (D-F) single colony isolation
Download PDF (422.6 KB)Acknowledgments
We thank Ms Tomoko Kojima for critical reading of the manuscript. This study was supported by Grants-in-Aid 22681010, 24117521, and 25440129 (to S.I.), and 21370015, 23120505, and 2424806 (to K.T.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a Sasakawa Scientific Research Grant from The Japan Science Society (to S.W.).
Notes
Abbreviations: GFP, green fluorescent protein; MA2 medium, modified Allen’s 2 medium.
References
- Kuroiwa T. BioEssays. 1998;20:344–354.
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, et al. Nature. 2004;428:653–657.
- Nozaki H, Takano H, Misumi O, Terasawa K, Matsuzaki M, et al. BMC Biol. 2007;5:28.
- Ohta N, Matsuzaki M, Misumi O, Miyagishima SY, Nozaki H, Tanaka K, Shin-I T, Kohara Y, Kuroiwa T. DNA Res. 2003;10:67–77.
- Ohta N, Sato N, Kuroiwa T. Nucleic Acids Res. 1998;26:5190–5198.
- Ohnuma M, Yokoyama T, Inouye T, Sekine Y, Tanaka K. Plant Cell Physiol. 2008;49:117–120.
- Watanabe S, Ohnuma M, Sato J, Yoshikawa H, Tanaka K. J. Gen. Appl. Microbiol. 2011;57:69–72.
- Ohnuma M, Misumi O, Fujiwara T, Watanabe S, Tanaka K, Kuroiwa T. Protoplasma. 2009;236:107–112.
- Imamura S, Kanesaki Y, Ohnuma M, Inouye T, Sekine Y, Fujiwara T, Kuroiwa T, Tanaka K. Proc. Natl. Acad. Sci. USA. 2009;106:12548–12553.
- Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Plant Cell Physiol. 2004;45:667–671.
- Ohnuma M, Kuroiwa T, Tanaka K. J. Gen. Appl. Microbiol. 2011;57:137–143.
- Niwa Y. Plant Biotechnol. 2003;20:1–11.
- Imamura S, Terashita M, Ohnuma M, Maruyama S, Minoda A, et al. Plant Cell Physiol. 2010;51:707–717.
