Abstract
A new bioactive compound, namely desmodianone H (1), and another known compound uncinanone B (2) were first isolated using bioactivity-guided isolation from the leaves of Lespedeza maximowiczii and structures were elucidated by comprehensive analysis of their nuclear magnetic resonance and mass spectrometry data. Compounds 1 and 2 exhibited strong inhibitory effects on mushroom tyrosinase activity.
Lespedeza maximowiczii (Leguminosae) is a deciduous shrub, which is generally distributed in both Eastern North America and Eastern Asia.Citation1) About 40 plant species belonging to the genus Lespedeza have been reported; these species show very diverse morphological characteristics.Citation2) Furthermore, many metabolites, especially isoflavanones,Citation3–6) phenolic compounds,Citation7), and chalconeCitation8) have been isolated from species belonging to this genus. Lespedeza plants have traditionally been considered to have antipyretic, anti-inflammatory, and diuretic activities.Citation9) In addition, compounds such as haginin A, dalbergioidin, and lespeflorin A–G that have been isolated from Lespedeza cyrtobotrya and Lespedeza floribunda are known to inhibit melanin synthesis in vivo.Citation10–12)
In our continuing research, we classified L. maximowiczii by significantly different phytochemicals between four species (L. cyrtobotrya, L. bicolor, L. cuneata, and L. maximowiczii), and L. maximowiczii showed the highest activity than other species (data not shown).Citation13) Herein, we report the isolation, structure elucidation, and tyrosinase inhibition activity of a new compound, desmodianone H (1), and a known compound, uncinanone B (2), from L. maximowiczii.
The leaves of L. maximowiczii were collected from Odo Mountain, Gajo Township, Guchang District, Gyeongsangnam-do, South Korea, in August 2011. The samples were identified by Joongku Lee et al., and a voucher specimen was deposited at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, Daejeon, Korea). Samples were dried under shade conditions and ground into a fine homogeneous powder by using a pulverizer. A sample of Lespedeza powder (120 g) was extracted with MeOH (3 × 600 mL, 2 d, room temperature, 200 rpm), and a methanol extract was obtained after evaporation of the solvent under vacuum, and it afforded 26% yield. The MeOH extract shows half-maximal inhibitory concentration value at 0.75 mg/mL. The extract was suspended in H2O and partitioned with EtOAc (1/1, v/v), and the crude EtOAc fraction shows the IC50 value at 0.12 mg/mL. The EtOAc fraction (2.98 g) was subjected to open column chromatography over Sephadex LH-20 resin (4 × 83 cm) by using isocratic elution with 80% MeOH to obtain 96 fractions.
These fractions were further purified with prep-HPLC using a YMC-Pack Pro C18 reversed-phase column (250 × 4.6 mm i.d. 5 μm; 5% CH3CN in H2O (v/v) – 100% CH3CN = 95:5–0:100, v/v at a flow rate of 1 mL/min) equipped with a Hitachi photodiode array detector to obtain compound 1 (tR 35 min, 7.8 mg) and compound 2 (tR 38 min, 19.2 mg). One-dimensional and 2D NMR spectra were measured in methanol-d4 (δH 3.30/δC 49.5) on a Bruker Avance 600 spectrometer (1H: 600 MHz, 13C: 150 MHz). HRESIMS was obtained from Waters micromass Q-TOF premier with UPLC Acquity System (Waters, Milford, MA, USA) and Varian 500-MS ion-trap mass spectrometer (Palo Alto, CA, USA) was used for mass fragment data. Optical rotation data were recorded with Jasco P-1020 polarimeter (Manasquan, New Jersey, USA). All the steps were selected on the basis of the mushroom tyrosinase inhibition activity assay, which is a basic method to confirm the inhibition of melanin synthesis.Citation14,15)
The mushroom tyrosinase inhibitory activity was determined as described previously with some modification.Citation11) The reaction mixture, consisted of 153 μL of 0.1 M sodium phosphate buffer (pH 6.5), 5 μL of sample dissolved in methanol, 5 μL of mushroom tyrosinase (2,500 unit/mL), and 36 μL of 1.5 mM L-tyrosine, was added to a 96 well plate. After the reaction mixture was incubated at 37 °C for 20 min, absorbance was measured at 490 nm with a microplate reader (Biotek, EL808, Seoul, Korea). The absorbance of the same mixture with the MeOH was used as control. Kojic acid was used as positive control. Each treatment was replicated three times. The percent inhibition of tyrosinase inhibition activity was calculated as follows: % Inhibition = [(C20 min – C0 min) – (S20 min – S0 min)/(C20 min – C0 min)] × 100, whereas C20 min is the absorbance of the control after 20 min, C0 min is the absorbance of the control after 0 min, S20 min is the absorbance of the experimental sample after 20 min and S0 min is the absorbance of the experimental sample after 0 min.
Compound 1 was obtained as a pale yellow powder, and its molecular formula was determined as C20H18O5 from its negative mode high resolution electron spray ionization mass spectroscopic data at m/z 337.1038 [M-H]− (calcd. For C20H17O5, 337.1076), which was compatible with the nuclear magnetic resonance data. The UV spectrum showed ketone absorption at λmax 239.0 nm; in addition, the bands at 298 and 330 (sh) nm were suggestive of a flavanone skeleton. The specific optical rotation of 1 had plus (+) sign ([α]d :+21 (c 0.07, MeOH)). The 13C NMR spectrum of 1 (Table ) displayed signals for 20 carbons, including one methyl group, one methylene, five methines, two methanetriyl groups, one carbonyl group, eight quarternary carbons, and two methylene bridges, as revealed by the heteronuclear single quantum coherence (HSQC) experiment. Interpretation of the 1H-1H correlation spectroscopy (COSY) data of 1 resulted in the identification of four proton spin-systems corresponding to H-2″–H-3″, H-5–H-6, H-2–H-3, and H-5′–H-6′ units, and the remaining connections were established by the analysis of heteronuclear multiple-bond correlations (HMBC). The HMBC correlations from H-2″ to C-7 and from H-3″ to C-8 and C-2″ were all connected as a dihydrofuran ring. In addition, the correlations from H-5 to C-7 and from H-6 to C-7 and C-8 led to the connection of C-8 with C-7, C-6, and C-5, which completed a benzofuran unit. The gross structure of 1 was determined by 1H-1H COSY, HSQC, and HMBC experiments, the key 1H-1H COSY and HMBC correlations being given in Fig. . The structure of 1 is similar to that of desmodianone G, a dihydroxy furanose flavanone isolated from Desmodium canum (which has a similar ring core skeleton).Citation16) However, desmodianone G has a hydroxylated quarternary carbon at C-5 and a quarternary carbon at C-6, with an extra methyl group. On the basis of the foregoing data, the structure of compound 1 was determined to be the new flavanone named desmodianone H.
Table 1. 1H (600 MHz) and 13C NMR (150 MHz) spectroscopic data (methanol-d4) of desmodianone H (1) and uncinanone B (2).
Fig. 1. Structure of compounds 1–2.
Notes: (A) Structures of compounds 1–2. (B) Key 1H→1H COSY (solid bond) and 1H→13C HMBC (arrow) correlations for 1.
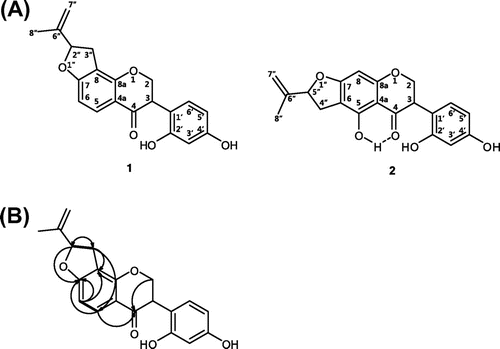
The compound 2 was identified as uncinanone B by comparing the physicochemical and spectroscopic data with previously reported data.Citation17) In the same way of the previous research, uncinanone B was detected as a diastereoisomers in our study. Uncinanone B has weak hydrogen bond between 5-OH and ketone group and this weak bond occurs at two signals with very small difference in NMR and UPLC-QTOF-MS data. To the best of our knowledge, this is the first report on the isolation of uncinanone B (2) from the leaves of L. maximowiczii.
The tyrosinase inhibition activity of the two isolated compounds, desmodianone H (1) and uncinanone B (2), was evaluated in vitro to calculate its IC50 values. Compounds 1 and 2 exhibited remarkably higher anti-tyrosinase activity, (IC50 values: 1.00 and 0.57 μm, respectively; Table ) compared to kojic acid (IC50 value, 84 μm), which was used as a positive control. Moreover, uncinanone B (2) showed higher potent inhibition rate than desmodianone H (1).
Table 2. Mushroom tyrosinase inhibitory activity of the bioactive compounds.
In conclusion, we isolated and identified the two compounds from L. maximowiczii leaves at the first time. Desmodianone H (1) is a novel compound that has tyrosinase inhibition activities. Together, uncinanone B (2) has higher anti-tyrosinase rate than desmodianone H. To our knowledge, this is the first report to investigate compounds from L. maximowiczii that inhibit the tyrosinase activities, suggesting that it could be a good candidate for skin-whitening agent.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.905180.
Supplemental Figs. 1-4
Download PDF (172.3 KB)Acknowledgements
This study was supported by the Bio-industry Technology Development Program (no. 110132-3), Ministry for Food, Agriculture, Forestry and Fisheries. This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ00952004).
Funding
This study was supported by the Bio-industry Technology Development Program (No. 110132-3), Ministry for Food, Agriculture, Forestry and Fisheries. This research was supported by a grant from the Eco-Innovation project of the Ministry of Environment, Korea [grant no. 416-111-006].
References
- Han JE, Chung KH, Nemoto T, Choi BH. Bot. J. Linn. Soc. 2010;164:221–235.10.1111/j.1095-8339.2010.01084.x
- Fant JB, Banai A, Havens K, Vitt P. Conserv. Genet. 2010;11:2195–2205.10.1007/s10592-010-0105-9
- Ueno A, Ichikawa M, Miyase T, Fukushima S, Saiki Y, Morinaga K. Chem. Pharm. Bull. 1973;21:1734–1740.10.1248/cpb.21.1734
- Ueno A, Ichikawa M, Fukushima S, Saiki Y, Noro T, Morinaga K, Kuwano H. Chem. Pharm. Bull. 1973;21:2715–2721.10.1248/cpb.21.2715
- Miyase T, Sano M, Yoshino K, Nonaka K. Phytochemistry. 1999;52:311–319.10.1016/S0031-9422(99)00194-6
- Maximov OB, Kulesh NI, Stepanenko LS, Dmitrenok PS. Fitoterapia. 2004;75:96–98.10.1016/j.fitote.2003.07.012
- Miyase T, Sano M, Nakai H, Muruoka M, Nakazawa M, Suzuki M, Yoshino K, Nishihara Y, Tanai J. Phytochemistry. 1999;52:303–310.10.1016/S0031-9422(99)00195-8
- Miyase T, Ueno A, Noro T, Fukushima S. Chem. Pharm. Bull. 1980;28:1172–1177.10.1248/cpb.28.1172
- Park HY, Kim GB, Kwon YS. Arch. Pharm. Res. 2010;33:1159–1163.10.1007/s12272-010-0804-2
- Kim JH, Beak SH, Kim DH, Choi TY, Yoon TJ, Hwang JS, Kim MR, Kwon HJ, Lee CH. J. Invest. Dermatol. 2008;128:1227–1235.10.1038/sj.jid.5701177
- Lee MY, Kim JH, Choi JN, Kim J, Hwang GS, Lee CH. J. Microbiol. Biotechnol. 2010;20:988–994.10.4014/jmb
- Mori-Hongo M, Takimoto H, Katagiri T, Kimura M, Ikeda Y, Miyase T. J. Nat. Prod. 2009;72:194–203.10.1021/np800395j
- Kim YM, Le J, Park S, Lee C, Lee JW, Lee D, Lee N, Lee D, Kim HY, and Lee CH. Plant Cell Rep. 2012;31:2085–2097.10.1007/s00299-012-1319-8
- Shamma M, Dudock SS. Tetrahedron Lett. 1965;43:3825–3828.10.1016/S0040-4039(01)89132-2
- Shamma M, Hillman MJ, Jones CD. Chem. Rev. 1969;69:779–784.10.1021/cr60262a002
- Zappia G, Menendez MP, Sampaio de Andrade Lima C, Botta B. Nat. Prod. Res. 2003;23:665–671.
- Tsanuo MK, Hassanli A, Hooper AM, Khan Z, Kaberia F, Pickett JA, Wadhams LJ. Phytochemistry. 2003;64:265–273.10.1016/S0031-9422(03)00324-8
