Abstract
The ppGpp-signaling system functions in plant chloroplasts. In bacteria, a negative effect of ppGpp on adenylosuccinate synthetase (AdSS) has been suggested. Our biochemical analysis also revealed rice AdSS homologs are apparently sensitive to ppGpp. However, further investigation clarified that this phenomenon is cancelled by the high substrate affinity to the enzymes, leading to a limited effect of ppGpp on adenylosuccinate synthesis.
Hyperphosphorylated guanine ribonucleotide, guanine-3′,5′-bispyrophosphate (ppGpp), is known as a global regulator in bacteria, and it regulates functions of multiple machineries in prokaryotic cell systems in response to various stress conditions.Citation1−4) The ppGpp-signaling system also occurs in plant chloroplasts, and it has been demonstrated that vascular plants have several distinct types of ppGpp synthetases. One of them, CRSH, is a plant-specific Ca2+-dependent ppGpp synthetase and its function is essential for plant reproduction.Citation5)
With respect to functions of ppGpp in plants, we have previously demonstrated that the chloroplast translation system is negatively regulated by high ppGpp concentration in vitro,Citation6) whilst another group demonstrated a physical interaction between the ppGpp analog with RNA polymerase, suggesting that chloroplast RNA polymerase is a possible target of ppGpp.Citation7) Regulatory roles of ppGpp in translation and transcription have already been demonstrated in bacteria,Citation3) and several lines of evidence have also suggested that adenylosuccinate synthetase (AdSS), a key enzyme in the purine biosynthetic pathway, is a potential ppGpp target in bacterial cells.Citation8) Substrates of AdSS are IMP, l-aspartate, and GTP, and these are converted to adenylosuccinate, GDP, and phosphate in the enzyme reaction. It has been known that plants have AdSS enzymes that are homologous to bacterial enzymes and localize into organelles.Citation9) Further, AdSS has been known as a target of a potent herbicide, hydantocidin, indicating an essential role of the enzyme in plants.Citation10) Here, we focused on the AdSS enzyme of rice, and characterized the effect of ppGpp on AdSS proteins based on kinetic parameter analysis by comparison with homologous enzymes from two model bacteria, Escherichia coli and Bacillus subtilis.
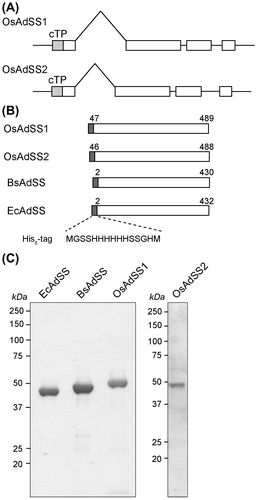
AdSS is a highly conserved enzyme among all living organisms, including vascular plants.Citation11) In addition to isolating an Arabidopsis thaliana homolog, we found two isoforms of AdSS in the rice genome database (http://ricexpro.dna.affrc.go.jp) which presumably localize into chloroplasts.Citation11) We designated these proteins as OsAdSS1 (accession No. NM_001057522) and OsAdSS2 (NM_001055663). We also cloned genes encoding AdSS from E. coli (EcAdSS, NC_007779) and B. subtilis (BsAdSS, NC_000964) for comparison. The expression vectors for these AdSS proteins were constructed as described below. The DNA segments of rice (Oryza sativa L. Nipponbare), organellar AdSS, OsAdSS1, and OsAdSS2 were obtained by 2-step PCR. First PCR for OsAdSS1 and OsAdSS2 was performed using cDNA prepared from O. sativaCitation12) and primer sets, OsAdSS1-for (5′-ATGCCATTCTCCCCTCCGTGCCTGGACCCC-3′) and OsAdSS1-rev (5′-CTATTTGTATATGAGAGCATCACGCCCTGGTCC-3′) for OsAdSS1 or OsAdSS2-for (5′-ATGTCGCTCTCCACTGTCAACCACGCCGCC-3′) and OsAdSS2-rev (5′-TTACTTGTATATTAGAGCATCCCTCCCAGGTCC-3′) for OsAdSS2. The amplified DNA fragments for each protein were used as templates for second-PCR with primer sets, OsAdSS1-for-2 (5′-TTTCCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCATATGTCCGTGGCCACGGCCCCGGA-3′) and OsAdSS1-rev-2 (5′-AAAGGATCCCTATTTGTATATGAGAGCATCACG-3′) for OsAdSS1 or OsAdSS2-for-2 (5′-TTTCCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCATATGGTGTCCGCGGCGGCTGTGGG-3′) and OsAdSS2-rev-2 (5′-AAAGGATCCTTACTTGTATATTAGAGCATCCCT-3′) for OsAdSS2. The protein-coding regions of BsAdSS and EcAdSS were amplified by colony-directed PCR with primer sets, BsAdSS-for (5′-TTTCCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCATATGTCTTCAGTAGTTGTAGTAGG-3′) and BsAdSS-rev (5′-AAAGGATCCTTAGTTCGCACGGTACAC-3′) for BsAdSS and EcAdSS-for (5′-TTTCCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCATATGGGTAACAACGTCGTC-3′) and EcAdSS-rev (5′-AAAGGATCCTTACGCGTCGAACGGGTC-3′) for EcAdSS, respectively. The amplified DNA fragments for each AdSS protein were cloned into pTA2 vector using Target Clone Kit (Toyobo, Osaka, Japan). These plasmids were digested with NcoI and BamHI, and the resultant DNA fragments for each AdSS gene were ligated into the pertinent cloning sites in pET19b (Novagen, Madison, WI).
All plasmids were transformed into E. coli BL21(DE3), and the transformed cells were grown at 37 °C in LB medium containing ampicillin at 100 μg mL−1. Expression of proteins was induced by the addition of 100 μM isopropyl β-d-1-thiogalactopyranoside into the culture when the cell density (OD600) reached 0.5. After continuous culture at 25 °C for 5 h, the cells were harvested by centrifugation and disrupted by sonication in a lysis buffer [50 mM disodium phosphate (pH 7.0), 0.5 M NaCl, 0.1% Triton X-100, 20% glycerol]. The cell extracts were centrifuged at 20,400× g for 15 min and the supernatants were loaded onto TALON metal affinity resin (BD bioscience, Franklin Lakes, NJ). After the resins were washed by the lysis buffer containing 18 mM imidazole, the proteins were eluted by the lysis buffer containing 180 mM imidazole. Concentrations of each purified protein were determined with Bio-Rad protein assay kit (Bio-Rad, Richmond, CA) using bovine serum albumin as a standard.
Activity for AdSS proteins was measured with spectrophotometer. The reaction mixture containing 20 mM HEPES-KOH (pH 7.7), 8 mM (CH3COO)2 Mg, 100 μM IMP, 50 μM GTP, and purified AdSS protein (100 nM protein for each) was pre-incubated at 30 °C for 5 min. After pre-incubation, the reaction mixtures were supplemented with 2 mM l-Asp (pH 7.7) for initiation of enzymatic reactions. The synthesis of adenylosuccinate was monitored as increase in absorbance at 280 nm with Bio-Spec-1600 spectrophotometer (Shimadzu, Kyoto, Japan). A unit of AdSS activity was defined as the formation of 1 μM of adenylosuccinate per min at 30 °C. The initial velocity of the reaction, Vo, was calculated from the slope of the linear portion. To assess the effects of ppGpp or GDP on the reaction, ppGpp (TriLink Biotechnologies, San Diego, CA) or GDP (Wako, Osaka, Japan) was added to the reaction mixture at the indicated concentration.
For OsAdSS1 and OsAdSS2, we predicted cleavage cites in N-terminal signal sequences based on sequence alignment. The AdSS from rice and bacteria was expressed as recombinant proteins in E. coli and purified to homogeneity (Fig. ). We observed that all four AdSS proteins exhibited catalytic activity and converted IMP to adenylosuccinate in vitro. Specific activities of enzymes were: 1.2 unit mg protein−1 for OsAdSS1, 0.75 for OsAdSS2, 0.60 for EcAdSS, and 0.81 for BsAdSS, respectively.
Fig. 1. Expression and purification of AdSS proteins.
Notes: (A) Genome structures of OsAdSS1 and OsAdSS2. Boxes and thin lines represent protein-coding sequences and introns, respectively, and cTP indicates an N-terminal chloroplast transit peptide. (B) Structures of recombinant AdSS proteins. Mature OsAdSS1 and OsAdSS2 proteins were designed based on the prediction of N-terminal chloroplast transit peptide, and were attached to a His-tag sequence, MGSSHHHHHHSSGHM, in each N-terminus for purification by immobilized metal affinity chromatography. Recombinant BsAdSS and EcAdSS proteins were also attached to the same N-terminal His-tag sequence. (C) Analysis of recombinant AdSS proteins. The purified proteins, 2.5 μg of proteins for EcAdSS, BsAdSS, and OsAdSS1, and 1 μg of protein for OsAdSS2, were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie Brilliant Blue staining.
We investigated the effects of ppGpp on catalysis of each AdSS protein. The catalytic activities of OsAdSS1 and OsAdSS2, as well as that of BsAdSS and EcAdSS were inhibited by ppGpp in a dose-dependent manner. We further examined the ppGpp inhibition of AdSSs and calculated kinetic parameters based on Lineweaver–Burk plot analysis (Fig. ). Results indicated that the inhibitory constant Ki for ppGpp against GTP substrate are 11 μM (OsAdSS1) and 6 μM (OsAdSS2), respectively (Table ). The kinetic parameters (Table ) also showed that affinities of substrate, GTP to OsAdSS1 ( = 9.1 μM), and OsAdSS2 (10 μM) are similar levels to that of ppGpp. On the other hand, compared to ppGpp and GTP, GDP, which is converted from GTP by the AdSS enzyme reaction, had more potent inhibitory effects on the activities of OsAdSS1 (
= 5.0 μM) and OsAdSS2 (6.7 μM) (Table ). Thus, our biochemical analysis revealed that the inhibitory effect of ppGpp on the plant AdSSs is limited by other guanine nucleotide concentrations.
Fig. 2. Lineweaver–Burk plots for recombinant AdSS proteins.
Notes: The activity of each AdSS protein (100 nM) was measured as described. The initial velocity of the reaction, Vo, was calculated from the slope of the linear portion at different concentrations of ppGpp/GDP (0–50 μM) over a range of GTP (10–50 μM).
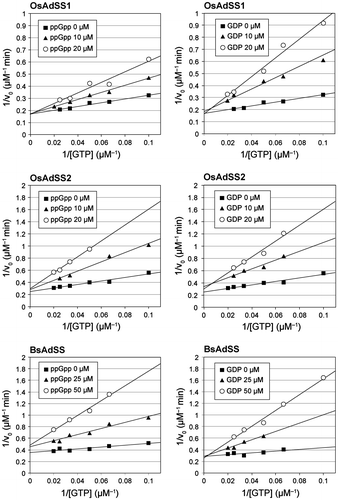
Table 1. Kinetic parameters of AdSS proteins.
The inhibitory effects of ppGpp on OsAdSS1 and OsAdSS2 seem to be more potent than that on EcAdSS ( = 50 μM), which had been reported as a ppGpp-sensitive enzyme previously,Citation8) whereas another bacterial counterpart, BsAdSS, exhibited ppGpp-sensitive activity in a similar range (
= 7.0 μM) of ppGpp concentrations observed in the examinations of rice enzymes (Table ). Thus, ppGpp sensitivities of rice AdSSs are likely more similar to B. subtilis AdSS than the E. coli homolog.
Based on further biochemical characterization of the bacterial counterparts, we revealed that BsAdSS shows significant sensitivity to GDP ( = 6.2 μM) and higher affinity to GTP (
= 4.4 μM) than that to ppGpp (
= 7.0 μM) (Table ). These results indicate that the inhibitory effect of ppGpp on BsAdSS, which may lead to reduction of ATP productivity, is also limited by concentrations of GTP and GDP. Further, it has been reported that the ATP level in B. subtilis is not affected by the elevation of cellular ppGpp concentration.Citation13) Although BsAdSS showed low Ki for ppGpp, which is within range of the cellular concentrations of ppGpp in B. subtilis, the apparent sensitivity of BsAdSS to ppGpp seems to be barely reflected in the ATP biosynthetic pathway in vivo.
Given the results from the characterization of rice AdSSs and BsAdSS, we hypothesized that ppGpp can affect AdSS catalytic activity only when ppGpp level markedly exceeds GTP level in the cells. In other words, it is normally difficult to compete out the substrate’s binding to the enzyme by ppGpp accumulation in vivo. The inhibitory effect of ppGpp on AdSS may terminate the cell system but it does not seem to be essential for cell survival.
In conclusion, we revealed that two AdSS enzymes from rice exhibit significant sensitivities to ppGpp in vitro, but precise biochemical analysis also clarified that these enzymes have similar affinity levels to GTP and ppGpp. The plant AdSS enzymes are possibly under the regulation of ppGpp, but the occurrence of the negative regulation seems to depend on quite limited conditions in vivo. It still remains difficult to quantify nucleotide levels of specific organelle due to the multiplicity of organelles in plant cells. Therefore, our biochemical data obtained in this study bring significant information to presume the regulation of AdSS enzymes in rice chloroplasts.
Acknowledgment
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology [grant-in-Aid for Scientific Research (number 24570054 to Y.T.)] and the Japan Society for the Promotion of Science [grant-in-Aid for JSPS Fellows to Y.N.].
Notes
Abbreviations: AdSS, adenylosuccinate synthetase; ppGpp, guanine-3′,5′-bispyrophosphate.
References
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, DC: ASM Press; 1996. p. 1458–1496.
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51.10.1146/annurev.micro.62.081307.162903
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008;11:100–105.10.1016/j.mib.2008.02.001
- Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21:174–180.10.1016/j.tim.2013.01.002
- Masuda S, Mizusawa K, Narisawa T, Tozawa Y, Ohta H, Takamiya K. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 2008;49:135–141.
- Nomura Y, Takabayashi T, Kuroda H, Yukawa Y, Sattasuk K, Akita M, Nozawa A, Tozawa Y. ppGpp inhibits peptide elongation cycle of chloroplast translation system in vitro. Plant Mol. Biol. 2012;78:185–196.10.1007/s11103-011-9858-x
- Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone guanosine 5-diphosphate 3-diphosphate (ppGpp) in plants. Proc. Nat. Acad. Sci. 2004;101:4320–4324.10.1073/pnas.0308555101
- Stayton MM, Fromm HJ. Guanosine 5-diphosphate 3-diphosphate inhibition of adenylosuccinate synthetase. J. Biol. Chem. 1979;254:2579–2581.
- Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006;57:805–836.10.1146/annurev.arplant.57.032905.105421
- Fonne-Pfister R, Chemla P, Ward E, Girardet M, Kreuz KE, Honzatko RB, Fromm HJ, Schar HP, Grutter MG, Cowan-Jacob SW. The mode of action and the structure of a herbicide in complex with its target: binding of activated hydantocidin to the feedback regulation site of adenylosuccinate synthetase. Proc. Nat. Acad. Sci. USA 1996;93:9431–9436.10.1073/pnas.93.18.9431
- Prade L, Cowan-Jacob SW, Chemla P, Potter S, Ward E, Fonne-Pfister. Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana. J. Mol. Biol. 2000;296:569–577.10.1006/jmbi.1999.3473
- Tozawa Y, Tanaka K, Takahashi H, Wakasa K. Nuclear encoding of a plastid sigma factor in rice and its tissue- and light-dependent expression. Nucleic Acids Res. 1998;26:415–419.10.1093/nar/26.2.415
- Tojo S, Satomura T, Kumamoto K, Hirooka K, Fujita Y. Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J. Bacteriol. 2008;190:6134–6147.10.1128/JB.00606-08
