Abstract
Kinesin light chain 1 (KLC1) mediates binding of KIF5 motor to specific cargo. Using the yeast two-hybrid screening, we found that mitochondrial fission protein dynamin-1-like protein (Dnm1L) interacted with KLC1, but not KIF5. Dnm1L and KLC1 were co-localized in cultured cells. These results suggest that KLC1 may play a potential role in post-fission mitochondrial transport.
Key words:
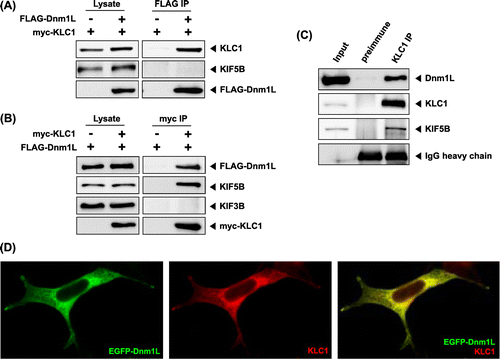
Kinesin 1 is a molecular motor protein that moves along microtubules to transport vesicles, organelles, protein complexes, and messenger RNAs in cells.Citation1) Kinesin 1 is a tetrameric protein composed of two heavy chains (KHCs) and two light chains (KLCs). The KHCs (KIF5A, KIF5B, and KIF5C) contain the motor domain, whereas KLCs (KLC1 and KLC2) are required for binding to cargo and also help keep the motor inactive.Citation1,2) The tetratricopeptide repeat (TPR) domain of KLCs is known as a protein–protein interaction motif, which consists of multiple tandem repeats of 34 amino acids.Citation3) The c-Jun NH2-terminal kinase (JNK)-interacting protein scaffold proteins were the first identified proteins to bind to kinesin 1 through the TPR domain.Citation4) Nearly 30 different cargoes moved by kinesin 1 have been identified, including amyloid precursor protein.Citation1,5,6) It remains to be investigated how kinesin 1 can bind to many different cargoes. In some cases, cargoes bind to adaptor/scaffolding proteins that mediate the attachment of kinesin 1 to cargo.Citation1) To improve the understanding of kinesin 1-dependent transport events, it was necessary to identify the interacting partners of kinesin 1. A yeast two-hybrid screening was performed with the Matchmaker LexA two-hybrid system (Clontech. Palo Alto, CA) using the mouse KLC1 fragment containing TPR domains (amino acids 80–510) as bait. From 1 × 10Citation7 clones of a mouse brain cDNA library subcloned into pB42AD vector, we obtained two clones containing cDNA fragments of dynamin-1-like protein (Dnm1L), also known as dynamin-related protein 1 (Drp1) (Fig. (A)). To identify the region of KLC1 required for interaction with Dnm1L, various fragments of KLC1 were constructed and tested for interaction with Dnm1L using yeast two-hybrid system (Fig. (B)). The result indicates that the region containing six TPR domains is required for binding. Dnm1L has highly conserved N-terminal three GTPase domains, a helical domain in the middle, and a GTPase effector domain (GED) at the C-terminal region.Citation7) The two clones contained the GED domain of Dnm1L. We investigated whether GTPase domains of Dnm1L also interacts with KLC1. As shown in Fig. (C), the fragment of Dnm1L containing three GTPase domains did not interact with KLC1. These results indicate that Dnm1L interacts with KLC1 through its GED domain. Next, we investigated whether KHCs (KIF5A, KIF5B, and KIF5C) and KIF3B, a motor subunit of kinesin 2, interact with Dnm1L. As shown in Fig. (A), KHCs and KIF3B did not interact with Dnm1L. GSK-3β, known to interact with Dnm1L, was used as a positive control.Citation8) A quantitative β-galactosidase assay also showed that Dnm1L is bound to KLC1 (Fig. (B)). The interaction between KLC1 and Dnm1L was further confirmed by GST pull-down using purified GST-Dnm1L and His-KLC1 (Fig. (C)). The result revealed a direct interaction between Dnm1L and KLC1. The KLC1–Dnm1L interaction was also assessed in mammalian cells. When the lysate of HEK-293T cells transiently expressing myc-KLC1 and FLAG-Dnm1L was immunoprecipitated with anti-FLAG antibody or anti-myc antibody, Dnm1L co-precipitated KLC1, but not KIF5B (Fig. (A)), and KLC1 co-precipitated both Dnm1L and KIF5B (Fig. (B)). These results indicate that Dnm1L interacts with free KLC1, but not with KLC1 bound to KIF5. In order to further confirm whether endogenous KLC1 and Dnm1L interact with each other, mouse brain lysates were subjected to immunoprecipitation with KLC1 antiserum. As shown in Fig. (C), Dnm1L was precipitated with KLC1. To determine whether KLC1 and Dnm1L co-localize in cells, KLC1 was co-expressed with EGFP-Dnm1L in HEK-293T cells. KLC1 and Dnm1L co-localized at the same region in cells (Fig. (D)). Taken together, these results indicate that Dnm1L is a new binding partner of KLC1.
Fig. 1. Identification of the proteins interacting with KLC1 by yeast two-hybrid screening.
Notes: (A) The domain structure of Dnm1L illustrating that positive clones possess a fragment of Dnm1L. GTPase and GED domains are indicated in gray. (B) Dnm1L-binding region in KLC1. The truncated forms of KLC1 were tested in the yeast two-hybrid assay for interaction with Dnm1L. +, interaction with Dnm1L; −, no interaction with Dnm1L. (C) Specific interaction of KLC1 with the GED domain of Dnm1L. Different truncations of Dnm1L were tested in the yeast two-hybrid assay for interaction with KLC1. +, interaction with KLC1; −, no interaction with KLC1; aa, amino acids.
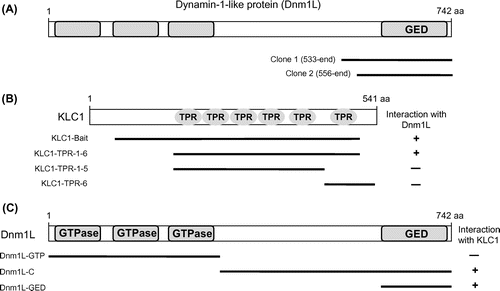
Fig. 2. Interaction of KLC1 or KIFs with Dnm1L.
Notes: (A) The C-terminal region of each protein was tested for interaction with full-length Dnm1L in yeast two-hybrid system. KLC1, but not KIFs, specifically interacted with Dnm1L. +, interaction with Dnm1L; −, no interaction with Dnm1L. (B) The strength of interactions of KLC1 or KIFs with Dnm1L was examined quantitatively using β-galactosidase activity in yeast two-hybrid reporter assay. (C) Direct binding of Dnm1L to KLC1 in GST pull-down assay using purified GST-fused Dnm1L and His-tagged KLC1.
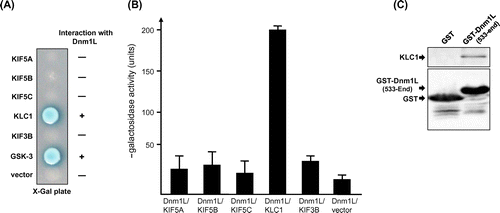
Fig. 3. Association of KLC1 with Dnm1L in co-immunoprecipitation and co-localization in mammalian cells.
Notes: (A & B) HEK-293T cells were transiently transfected with myc-KLC1 and FLAG-Dnm1L plasmids as indicated. Cell lysates prepared using lysis buffer with 0.5% NP-40 were immunoprecipitated with anti-FLAG antibody in (A) or anti-myc antibody in (B). Precipitates were immunoblotted with anti-KLC1, KIF5B, KIF3B, or FLAG antibodies. Dnm1L specifically co-precipitated KLC1 but not KIF5B nor KIF3B. (C) Mouse brain lysates were immunoprecipitated with anti-KLC1 antibody or preimmune serum, and then the precipitates were immunoblotted with anti-Dnm1L, KLC1, or KIF5B antibodies. (B) Twenty-four hours after transfection with KLC1 and EGFP-Dnm1L plasmids, HEK-293T cells were subjected to immunofluorescence staining with anti-KLC1 antibody.
Mitochondria are dynamic organelles that undergo trafficking, fusion, and fission. Dnm1L is required for mitochondrial fissionCitation9–11) and for the correct distribution of mitochondria.Citation12) Mutation in Drp1, the Drosophila homolog of Dnm1L, leads to abnormal clustering of mitochondria in neuronal cell bodies, causing dramatic defection in synaptic localization of mitochondria.Citation12) Mitochondrial fission by Dnm1L may relate to the availability of small transportable mitochondria, which is important for proper transport of mitochondria to synaptic terminals.Citation13) Axonal transport of mitochondria is directly mediated by KLC-independent interaction between milton and KIF5.Citation14,15) Milton acts as an adaptor protein that links KIF5 motor to mitochondria.Citation15) In mutants lacking milton, mitochondria stay in neuronal cell bodies and do not move to synaptic terminals.Citation14) KLC1 inhibits the interaction between KIF5 and milton.Citation15) In this study, Dnm1L is found to interact with free KLC1, but not with KIF5-bound KLC1 (i.e. the KLC1 as a subunit of kinesin 1). Taken together, we speculate that, after mitochondrial fission, interaction of Dnm1L with KLC1 may lead to dissociation of kinesin 1 tetramer, allowing KIF5 to interact with milton and transport mitochondria.
Additional information
Funding
References
- Hirokawa N, Niwa S, Tanaka Y. Molecular motor in neuron: mechanism and roles in brain function, development, and disease. Neuron. 2010;68:610–638.10.1016/j.neuron.2010.09.039
- Rahman A, Friedman DS, Goldstein LS. Two kinesin light chain genes in mice. Identification and characterization of the encoded proteins. J. Biol. Chem. 1998;273:15395–15403.10.1074/jbc.273.25.15395
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939.10.1002/(SICI)1521-1878(199911)21:11<>1.0.CO;2-8
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 2001;152:959–970.10.1083/jcb.152.5.959
- Gindhart JG. Towards an understanding of kinesin-1 dependent transport pathways through the study of protein–protein interactions. Brief. Funct. Genomic. Proteomic. 2006;5:74–86.10.1093/bfgp/ell002
- Muresan Z, Muresan V. Coordinated transport of phosphorylated amyloid-β precursor protein and c-Jun NH2-terminal kinase-interacting protein-1. J. Cell Biol. 2005;171:615–625.10.1083/jcb.200502043
- Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Rev. 2011;67:103–118.10.1016/j.brainresrev.2010.11.004
- Hong YR, Chen CH, Cheng DS, Howng SL, Chow CC. Human dynamin-like protein interacts with the glycogen synthase kinase 3beta. Biochem. Biophys. Res. Commun. 1998;249:697–703.10.1006/bbrc.1998.9253
- Otera H, Ishihara N, Mihara K. New insights into the function and regulation of mitochondrial fission. BBA Mol. Cell Res. 2013;1833:1256–1268.
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256.10.1091/mbc.12.8.2245
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536.10.1146/annurev.genet.38.072902.093019
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378.10.1016/j.neuron.2005.06.018
- Aldridge AC, Benson LP, Siegenthaler MM, Whigham BT, Stowers RS, Hales KG. Roles for Drp1, a dynamin-related protein, and milton, a kinesin-associated protein, in mitochondrial segregation, unfurling and elongation during Drosophila spermatogenesis. Fly. 2007;1:38–46.10.4161/fly
- Stowers RS, Megeath LJ, GorskaAndrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077.10.1016/S0896-6273(02)01094-2
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 2006;173:545–557.10.1083/jcb.200601067
