Abstract
We previously described potential probiotic Lactobacillus rhamnosus strains, isolated from fermented mare milk produced in Sumbawa Island, Indonesia, which showed high adhesion to porcine colonic mucin (PCM) and extracellular matrix (ECM) proteins. Recently, mucus-binding factor (MBF) was found in the GG strain of L. rhamnosus as a mucin-binding protein. In this study, we assessed the ability of recombinant MBF protein from the FSMM22 strain, one of the isolates of L. rhamnosus from fermented Sumbawa mare milk, to adhere to PCM and ECM proteins by overlay dot blot and Biacore assays. MBF bound to PCM, laminin, collagen IV, and fibronectin with submicromolar dissociation constants. Adhesion of the FSMM22 mbf mutant strain to PCM and ECM proteins was significantly less than that of the wild-type strain. Collectively, these results suggested that MBF contribute to L. rhamnosus host colonization via mucin and ECM protein binding.
Graphical Abstract
Binding of MBF from L. rhamnosus FSMM22 to Mucin and ECM proteins.

Lactic acid bacteria of the genus Lactobacillus comprise one of the most important health-promoting bacterial groups in the human intestinal microbiota.Citation1) Adhesion to the intestinal mucosal surface through mucins and a variety of extracellular matrix (ECM) proteins is an important prerequisite for the colonization of Lactobacillus in the intestinal lumen, providing them a competitive advantage in this ecosystem.Citation2,3) Lactobacillus rhamnosus is frequently isolated from the human gastrointestinal (GI) tract and dairy products.Citation4,5) The GG strain of L. rhamnosus (LGG) is a well-established probiotic strain. The health-benefiting properties of this strain are partially dependent on its prolonged residence in the GI tract, and they are probably influenced by its adhesion to the intestinal mucosa.Citation6) The LGG strain displays specialized surface adhesions; for example, the pilin SpaC subunit, located within the Spa pili structure, binds to human mucinCitation7,8) and intestinal epithelial cells.Citation9) MabA is an LPXTG cell wall-anchoring protein, which modulates adhesion to epithelial cells and biofilm formation.Citation10)
Recently, an internalin J adhesin homolog, mucus-binding factor (MBF, LGG_02337), was discovered in LGG.Citation11) Although recombinant MBF protein binds immobilized human mucins, no significant differences were observed between the wild-type and mbf mutant strains in mucin adhesion ability.Citation9,11) Therefore, the MBF protein is presumed to play an ancillary role in pilus-mediated mucosal adhesion by LGG. The MBF protein contains a four-repeat Pfam cell wall surface anchor repeat (PF13461). The cell wall surface anchor repeat occurs in several cell wall surface proteins in Listeria monocytogenes.Citation12) These proteins attach to the L. monocytogenes surface and have pleiotropic functions, including peptidoglycan metabolism, protein processing, mucosal surface adhesion, and host tissue invasion.Citation13–15) Moreover, the cell wall surface anchor repeat is a member of clan Gram-pos_anchor (CL0501), which contains mucin-binding protein domain (MucBD, PF06458). The MucBD-containing proteins have been predominantly identified in lactobacilli that are naturally found in intestinal nichesCitation16) and promote cell adhesion to mucins.Citation17–19) However, the cell wall surface anchor-containing MBF adhesion properties to intestinal mucosa-associated components, including ECM proteins, are poorly studied in L. rhamnosus.
We previously described the properties of 25 potential probiotic L. rhamnosus strains isolated from traditional Indonesian foods, including fermented Sumbawa mare milk (FSMM) produced on Sumbawa Island.Citation20) All isolates showed resistance to bile salts and acidity. Interestingly, three L. rhamnosus strains, FSMM 15, 22, and 26, exhibited high adhesion to porcine colonic mucin (PCM) and ECM proteins. Moreover, FSMM22 showed significantly higher adhesion to laminin than the LGG strain. Here, we report binding characteristics of MBF protein to mucins and ECM proteins as determined by surface plasmon resonance (SPR) using a Biacore X Instrument. We also present findings of MBF protein function in promoting FSMM22 adhesion.
Materials and methods
Bacterial strains and growth conditions
LGG and 14 FSMM strainsCitation20) were cultured on de Man–Rogosa–Sharpe (MRS) agar plates (BD Difco, Le Pont de Claix, France) at 28 °C or 37 °C under anaerobic conditions. Escherichia coli strains DH5α and Rosetta2 (Stratagene, La Jolla, CA, USA) were grown in Luria–Bertani (LB) broth or on LB agar plates at 37 °C. Lactococcus lactis subsp. lactis IL1403 was grown in M17 medium (BD Difco) supplemented with 0.5% (w/v) glucose at 30 °C. Ampicillin (100 μg/mL), kanamycin (50 μg/mL), chloramphenicol (30 μg/mL), and erythromycin (10 μg/mL) were added when necessary.
Molecular cloning of the mbf gene from L. Rhamnosus FSMM22
DNA regions upstream and downstream of the mbf gene (approx. 1800 bp) were amplified by PCR using Ex Taq DNA polymerase (Takara Bio, Shiga, Japan) using FSMM22 genomic DNA as template and primer pair S1/S2 (Table ). The amplified fragments were inserted into the pGEM-T Easy vector (Promega, Tokyo, Japan) to obtain pGEM-T-mbf. The resulting plasmids were sequenced. The nucleotide sequence of the mbf gene from FSMM22 was deposited in GenBank under the accession number AB968049. The signal peptide and transmembrane domain were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), respectively.
Table 1. Primers used in this study.
Construction and expression of 6×Histidine tag fused–MBF
The expression vector pET28b (Novagen, Madison, WI, USA) was designed to express recombinant MBF fused to a 6×Histidine tag at the C terminus (His6-MBF). The mbf gene, without the region encoding the N-terminal secretion signal or the C-terminal sortase recognition site, was PCR amplified from LGG or FSMM22 genomic DNA using the gene-specific primers S14 (for LGG, forward), S19 (for FSMM22, forward), and S15 (for LGG and FSMM22, reverse) (Table ). The PCR products were digested with NcoI and XhoI restriction endonucleases and ligated into a similarly digested expression vector, pET28b. Each plasmid construct was transformed into E. coli DH5α cells. The resulting plasmids were confirmed by sequencing, and all plasmid DNAs were introduced into E. coli Rosetta2 cells. Transformed cells were grown in LB medium at 37 °C with shaking. When the OD600 reached 0.5, isopropyl-β-d-thiogalactopyranoside (0.1 mM) was added to induce protein expression. After cultivation at 37 °C for 4 h, the cells were harvested and lysed in BugBuster Protein Extraction Reagent (Novagen) to obtain cell-free extracts. His6-MBF was purified by Ni2+-nitrilotriacetic acid affinity chromatography and ion exchange chromatography using HisTrap and SP-Sepharose FF columns, respectively (GE Healthcare). The protein fractions of interest were pooled and dialyzed against 10 mM HEPES buffer (pH 7.2). Protein purity was assessed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5% polyacrylamide), and concentrations were determined spectrophotometrically by the BCA method (Thermo Scientific, Wilmington, DE, USA).
Construction of FSMM22 mbf-deletion mutant
The primer pairs S1/S3 and S4/S2 (Table ) were used to introduce restriction sites in the middle of the FSMM22 mbf gene by PCR. The amplified fragment was inserted into the pGEM-T Easy vector. The resulting plasmid was digested with BamHI and HindIII. The chloramphenicol resistance cassette from pGK12Citation21) was amplified using the 1094/1934 primer pair (Table ) and digested with BamHI. These three fragments were ligated to the BamHI site in the middle of the mbf gene. The resultant fragment was amplified using the S1/S2 primer pair and inserted into the pGEM-T Easy vector, designated as pGEM-T-mbf::cmr. This plasmid was digested with EcoRI and HindIII and was subsequently ligated into the temperature-sensitive cloning vector pG+host6 (Appligene, Pleasanton, CA, USA). The resulting plasmid was then electroporated (1.7 kV, 200 Ω, 2.5 μF) into FSMM22 cells, as previously described.Citation22) Following 2 days of incubation at 28 °C, transformants were selected by chloramphenicol resistance and erythromycin sensitivity, as previously described.Citation23) To determine whether the resultant construct pG+host6 mbf::cmr marker, which was inserted into the genome, encoded for the mbf mutant gene, a double-crossover recombination was confirmed by PCR using the S1/S2 primer pair. The PCR products were analyzed by 1% (w/v) agarose gel electrophoresis.
To complement the FSMM22 mbf-deletion mutant, pGEM-T-mbf was digested with EcoRI, and then the fragment was cloned into the EcoRI restriction sites of pIL253,Citation24) designated as pIL253-mbf. This plasmid was electroporated into L. lactis IL1403, according to a previously described method.Citation25) The resulting plasmid was electroporated into the FSMM22 mbf-deletion mutant strain.
Overlay dot blot analysis
Ten micrograms of mouse laminin (BD Biosciences), human fibronectin (Sigma-Aldrich, St. Louis, MO, USA), human collagen IV (Sigma-Aldrich), bovine serum albumin (BSA; Sigma-Aldrich), or 100 ng of PCM as the hexose equivalentCitation26) were immobilized onto nitrocellulose membranes (Whatman, Kent, UK). Membranes were blocked in HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20; pH 7.2) containing 5% (w/v) skim milk for 2 h, followed by incubation with His6-MBFs (500 nM) in HBS-EP containing 1% (w/v) skim milk for 3 h at room temperature. After washing three times with HBS-EP, the membranes were incubated with anti-His tag antibody (Novagen, 1:1500 dilution) for 1 h. After washing, the membranes were exposed to alkaline phosphatase-conjugated anti-mouse IgG (Dako; 1:2500 dilution) in HBS-EP at room temperature for 1 h. The blots were developed using a BCIP/NBT liquid substrate system (Sigma-Aldrich).
SPR analysis
Binding affinity of MBF for mucin and ECM proteins was assessed by SPR on a Biacore X Instrument (GE Healthcare, NJ, USA). Laminin, fibronectin, collagen IV, PCM, and BSA were immobilized on a CM5 dextran sensor chip (GE Healthcare) with 4665, 6681, 5242, 4531, and 3130 resonance units (RUs), respectively, using amine-coupling reagents (GE Healthcare). The binding of His6-MBFs to the coated surface was determined using HBS-EP buffer (pH 7.2) with Biacore X at a flow rate of 20 μL/min. Concentrations of the analytes are indicated in Fig. (b). The dissociation step was performed at the same flow rate for 3 min. The signal from each binding experiment was corrected for nonspecific binding by subtracting the signal obtained from the blank surface. Finally, regeneration of the sensor surface was achieved through a 60-s exposure to 50 mM tris–HCl containing 2 M NaCl (pH 9.5). The association rate (ka), dissociation rate (kd), and dissociation constant (KD = kd/ka) were calculated using the BIA Evaluation Software version 3.0 (GE Healthcare). Global analysis was performed using the simple 1:1 Langmuir binding model.
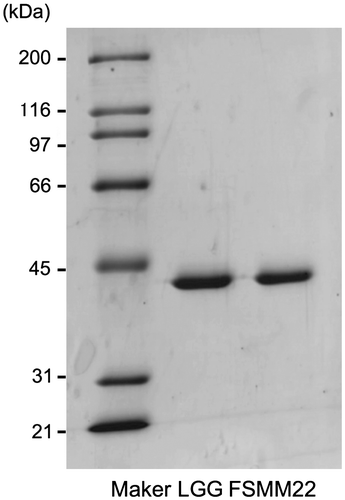
Fig. 1. Production and purification of His6-MBF.
Notes: Purified recombinant His6-MBFs from LGG and FSMM22 were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Molecular mass standards are indicated on the left.
To investigate the effect of ionic strength on MBF-PCM and MBF-ECM proteins interactions, HBS-EP buffer was supplemented with NaCl to a final concentration ranging from 150 to 450 mM. His6-MBF (500 nM) was injected over immobilized His6-MBF. RU values were measured at the end of dissociations without further sample addition. For further characterization of MBF self-association, His6-MBF was immobilized on a CM5 dextran sensor chip with 2543 RU. His6-MBF (1000 nM) was injected over immobilized His6-MBF. The same experimental step as described above was performed.
Preparation of cell surface proteins
The LGG and 14 FSMM strains were cultivated anaerobically in 30 mL of MRS broth at 37 °C for 10 h. Cell surface proteins were extracted as previously describedCitation11) with the following modifications: bacteria were harvested by centrifugation (6000 × g, 10 min, 4 °C), and the pelleted cells were then rinsed once with phosphate-buffered saline (PBS). Cells were resuspended in 200 μL of extraction buffer (50 mM tris–HCl pH 8.0, 30% [w/v] sucrose, 5 mM MgCl2, 5 mM CaCl2, 4 mg/mL lysozyme, and 150 U/mL mutanolysin) and then incubated at 37 °C for 3 h, followed by an additional centrifugation at 16,000 × g for 10 min at 4 °C. The supernatants were collected for western blotting. Whole-cell lysates were suspended in 300 μL of 50 mM tris–HCl (pH 8.0) and 0.3 g of 0.1 mm zirconia–silica beads. Total suspension was achieved by beating for 180 s at 4800 rpm in a bead beater (FastPrep QBiogene, Carlsbad, CA, USA). The debris was removed by centrifugation at 10,000 × g for 5 min at 4 °C. The supernatants were collected for Western blotting.
Western blotting
Antisera against the MBF synthetic peptide “CRYVRLAADSAAASGTFPKD” were raised in rabbits by routine immunization procedures. Twenty micrograms of protein samples were separated by SDS-PAGE (12.5% polyacrylamide) and transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were blocked with 5% (w/v) skim milk in PBS-0.05% Tween 20 (PBS-T) for 3 h at room temperature. After washing membranes with PBS-T, anti-MBF antibodies (diluted 1:300 in PBS-T) were added and incubated for 1 h at room temperature. Membranes were then washed and incubated for 1 h with horseradish peroxidase-conjugated mouse anti-rabbit IgG (Sigma-Aldrich) diluted at 1:1500 in PBS-T. After washing, the signal was developed with a TMB membrane peroxidase substrate (KPL, Gaithersburg, MD, USA).
Bacterial adhesion assay
A bacterial adhesion assay was conducted as previously described,Citation20) with some modifications. A 96-well microplate was coated with PCM, fibronectin, laminin, collagen IV, and BSA (negative control). L. rhamnosus strains were cultivated in MRS broth at 37 °C until the OD600 reached 1.0. Cells were harvested by centrifugation (6,000 × g, 5 min, 4 °C) and suspended in Dulbecco’s Modified Eagle’s Medium (DMEM). The bacterial suspension was added to each well and incubated for 1 h at 37 °C. After washing twice, 100 μL of 0.01% Triton X-100 in DMEM were added to each well, and the bacterial cells were suspended by vigorous pipetting. Serial dilutions of suspended bacteria were plated on MRS agar. Adhesion results were expressed as percentages calculated from three independent experiments, as follows: 100× (number of adhering bacteria/number of bacteria inoculated).
Statistical analyses
PRISM6 software (GraphPad Software) was used for all statistical analyses. Significant differences were determined using one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test. “n” represents the number of individual experiments. Differences with p-values of less than 0.05 were considered statistically significant. In SPR analysis, the chi-squared values of all data were calculated using version 3.0 BIA Evaluation Software.
Results
Cloning and sequence analysis of mbf gene from strain FSMM22
Cloning and sequence analyses showed that the FSMM22 mbf gene consists of a 1317-bp open reading frame (AB968049) encoding a polypeptide of 438 amino acids with a predicted molecular mass of 46.4 kDa, including a putative N-terminal secretion signal peptide (residues 1–39) and an “LPNTN” cell wall anchor domain (residues 402–406) containing a C-terminal transmembrane region (residues 402–438). MBF of the FSMM22 strain shares 97% identity with that of the LGG protein (YP_003172083.1).
Binding properties of MBF to mucus components
The recombinant His6-MBF from FSMM22, expressed in E. coli Rosetta2 cells and purified on HisTrap and ion exchange columns, produced a single protein band with a molecular mass of approx. 40 kDa, as determined by SDS-PAGE (Fig. ). The recombinant His6-MBF from LGG was also prepared to compare its property with MBF from FSMM22.
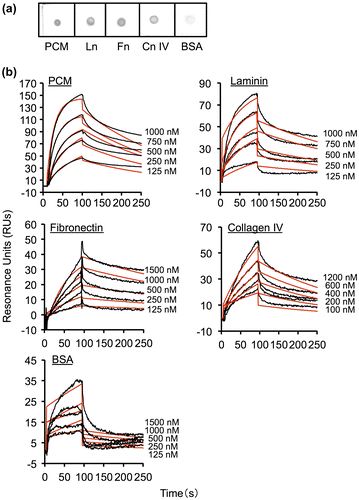
The binding properties of MBF from FSMM22 to PCM and ECM proteins were determined by overlay dot blot assays. PCM and ECM proteins were blotted onto a membrane, then His6-MBF from FSMM22 was overlaid. His6-MBF showed strong binding to PCM, laminin, collagen IV, and fibronectin (Fig. (a)). In contrast, His6-MBF exhibited very limited binding to BSA. Binding of His6-MBF to PCM and ECM proteins was also evaluated by SPR. As shown in Fig. (b), the interactions of His6-MBF from FSMM22 were dose-dependent and saturable. The experimental data showed a good fit to the calculated curves, suggesting that the model adequately describes the data. From the kinetic quantitative data for these interactions, the KD was determined to be 5.8 × 10−8 M for the interaction with PCM, 1.9 × 10−7 M for the interaction with laminin, 1.9 × 10−7 M for the interaction with fibronectin, and 1.6 × 10−7 M for the interaction with collagen IV (Table ). Moreover, the KD value for the binding of His6-MBF to BSA was determined to be 1.5 × 10−5 M. These data indicate that in addition to binding to PCM, FSMM22 MBF binds to several ECM proteins. The KD value for BSA is approx. 2 log-fold higher than that for PCM and ECM proteins. In addition, His6-MBF from LGG showed similar patterns of binding to PCM and ECM proteins (Table S1). To analyze MBF self-associates, we examined the binding of MBF to MBF immobilized on a biosensor. MBF showed little or no self-association (Fig. S1).
Fig. 2. Binding of His6-MBF from FSMM22 to PCM and ECM proteins.
Notes: (a) Overlay dot blot showing binding of His6-MBF to PCM, laminin (Ln), fibronectin (Fn), collagen IV (Cn IV), and BSA. Bound PCM and ECM proteins were detected using an anti-His tag antibody. (b) Biacore X sensorgrams of the interaction of His6-MBF with PCM, laminin, fibronectin, collagen IV, and BSA. His6-MBFs at the indicated concentrations were injected onto a CM5 sensor chip with immobilized PCM or ECM proteins. The measured data (black line) and their global fits are overlaid (red line). The KD value is indicated in the text and Table
Table 2. Affinity and rate constants for interactions of His6-MBF from FSMM22 with PCM and ECM proteins determined by SPR analysis.
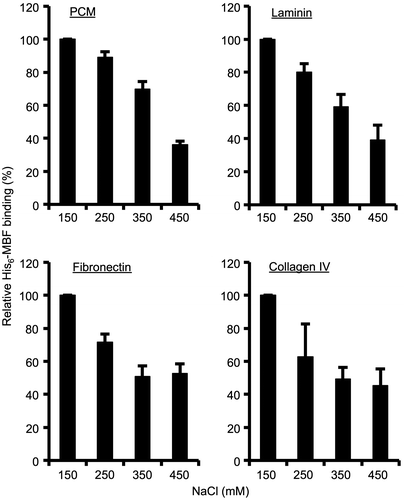
To determine whether the interaction between MBF and PCM or ECM is ionic, binding assays were conducted in the presence of varying NaCl concentrations. Binding of MBF to PCM, laminin, fibronectin, and collagen IV decreased with increasing NaCl concentrations (Fig. ). Thus, MBF binding to PCM and ECM proteins is influenced by ionic strength.
Fig. 3. Effect of different NaCl concentrations on His6-MBF binding.
Notes: Binding of His6-MBF (500 nM) from FSMM22 to immobilized PCM and ECM proteins was analyzed by SPR. HBS-EP buffer was supplemented with NaCl to a final concentration ranging from 150–450 mM. Results were expressed as relative percentage His6-MBF binding compared to 150 mM NaCl. Error bars indicate SD (n = 3).
Characterization of the mbf mutant of strain FSMM22
Adhesion assays were performed to determine the contribution of MBF to FSMM22 cell adhesion to PCM and ECM proteins, an FSMM22 mbf mutant was accordingly generated. The resultant cmr gene construct (approx. 850 bp), which incorporated into the genome by double-crossover recombination and encoded the mbf mutant gene, was confirmed by PCR (Fig. (a)). In addition, the absence of MBF protein was confirmed by Western blotting of whole-cell lysates with an anti-MBF antibody. A band of approx. 45 kDa was readily observed in the wild-type strain, but not in the mbf mutant strain (Fig. (b)). In contrast, the band was detected when the mbf mutant was complemented with pIL253-mbf (i.e. mbf-complemented strain).
Fig. 4. Effect of mbf deletion on FSMM22 adhesion to PCM and ECM proteins.
Notes: (a) A genotype analysis of the resultant cmr gene into the genome-encoded mbf gene was performed by PCR using primer pair S1/S2. Amplified fragments were subjected to agarose gel electrophoresis. Lanes: 1, size maker; 2, wild-type strain; and 3, mbf mutant. The sizes of representative marker fragments are shown to the left (kb). (b) MBF was detected in the whole-cell lysates of FSMM22 wild-type strain (Lane 1), mbf mutant (Lane 2), and mbf-complemented strain (Lane 3) by Western blotting with an anti-MBF antibody. The position of the MBF protein is highlighted (arrow). The sizes of representative marker fragments are shown to the left (kDa). (c) Adhesion was examined for FSMM22 wild-type strain (black bar), mbf mutant (white bar), and mbf-complemented strain (gray bar) to PCM, laminin, fibronectin, collagen IV, and BSA. Asterisks indicate significant differences in binding (*p < 0.05) compared to wild-type strain, as analyzed by one-way ANOVA with the Dunnett’s post hoc test. (n = 5).
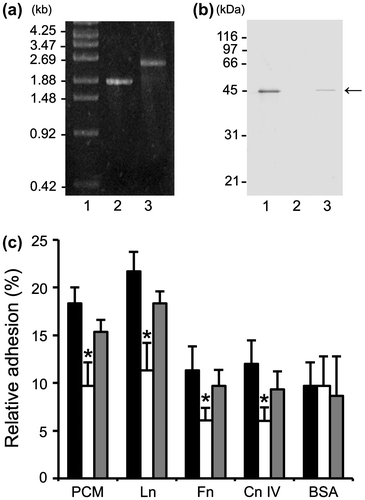
Subsequently, the effect of the mbf deletion on cellular adhesion to PCM and ECM proteins was examined. Adhesion of mbf mutants to PCM, laminin, collagen IV, and fibronectin decreased significantly, compared to the wild type (Fig. (c)). No significant differences were observed between the wild-type strain and the mbf mutant in adhesion to BSA. Although a small decrease was observed in adhesion of the mbf-complemented strain to PCM and ECM proteins, these differences were not statistically significant when compared with the wild-type strain. These results implicate MBF in L. rhamnosus FSMM22 adhesion to mucin and several ECM proteins.
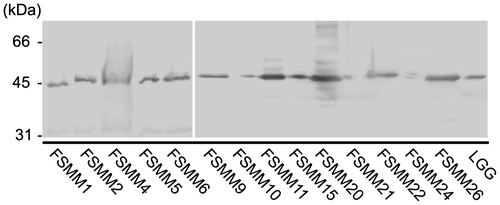
Detection of MBF in cell wall surface extracts of the LGG and FSMM strains
Western blot analysis was used to determine the cell surface expression levels of MBF protein in the LGG and 14 FSMM strains (Fig. ). MBF was detected in cell wall surface extracts of all strains, although at lower levels in the FSMM 10, 21, and 24 strains, suggesting that there are different expression levels at the cell surface or reduced cross-reaction with the anti-MBF antibody.
Discussion
In this study, we demonstrated that His6-MBF from FSMM22 binds to PCM, laminin, collagen IV, and fibronectin. In addition, adhesion of the FSMM22 mbf mutant strain to PCM and ECM proteins was significantly reduced compared with that of the wild-type strain. Involvement of LGG MabA has been speculated in the adhesion of L. rhamnosus to ECM, but no significant differences were observed between the wild-type and mabA mutant strains.Citation10) Moreover, the SpaC pili subunit of LGG was shown, by atomic force microscopic analysis, to interact with collagen.Citation27) However, the involvement of SpaC pili in LGG adhesion processes has not been examined. Therefore, factors adhering to ECM proteins in L. rhamnosus have yet to be determined. To our knowledge, this is the first detailed study characterizing an ECM adhesion factor in L. rhamnosus. The present study also indicated that MBF has the ability to bind to several mucus components. MBF possibly contributes to produce the strong avidity of bacterial cells for intestinal mucosal surfaces.
Our kinetic analysis showed that the interactions of His6-MBF with several ECM proteins are dose-dependent, saturable, and have submicromolar KD values. Furthermore, compared with PCM and ECM proteins, the binding of MBF to BSA exhibited high KD values, and did not exhibit self-association. By contrast, the MBF binding is shown to be dose-dependent and influenced by ionic strength. On the basis of these results, we speculate that electrostatic interactions are partially responsible for MBF binding in a specific recognition process. Similar interactions have been found in endopeptidase O (PepO) from Streptococcus pneumonia,Citation28) and serine-aspartate repeat F from Staphylococcus epidermis.Citation29) The binding of these proteins to ECM proteins was disrupted in the presence of salt and was also affected by pH, cation concentration, detergents, and basic amino acids. These studies suggested that the adhesive potential of bacteria depends on both cell surface hydrophobicity and ionic strength. Additional studies are needed to determine whether MBF binding is also mediated by both hydrophobic or other receptor-ligand interactions and electrostatic interactions.
Using bacterial adhesion assays, we demonstrated that MBF plays an important role in FSMM22 adhesion to PCM and ECM proteins. In addition, there was no difference in the adhesion of FSMM22 to BSA in the wild-type strain and the mbf mutant, indicating that the low affinity between MBF and BSA (KD = 1.5 × 10−5 M) might not contribute to bacterial adhesion. In L. rhamnosus, MBF protein was found in all tested FSMM strains, suggesting that it is conserved among the strains. Moreover, within FSMM22 and LGG, both His6-MBFs could bind to PCM and ECM proteins. Interestingly, a previous study showed that the adhesion capacity of the LGG mbf mutant strain was not abrogated compared with the wild-type strain.Citation9,11) Although the expression of MBF protein, as indicated by Western blotting, in the FSMM10, 21, and 24 strains was much lower than in other strains, adhesion to PCM and ECM proteins was moderate among 14 FSMM strains, which contradicts previously described adhesion properties.Citation20) Taken together, our results and those of previous studies suggest that, although MBF appears to contribute significantly toward the binding of L. rhamnosus to PCM and ECM proteins, the adhesion properties are probably specific to particular L. rhamnosus strains. Moreover, the adhesion of LGG involving MBF is thought to be influenced by extracellular polysaccharides and pili.Citation11) Recently, an extensive genomic analysis of the L. rhamnosus species demonstrated that the production of functional Spa pili is significantly more prevalent in human isolates than in dairy product isolates.Citation5) Thus, further analysis is required to confirm the localization of an extracellular polysaccharide and pilus on the cell surface of FSMM strains.
To summarize, we have provided experimental evidence demonstrating that MBF promotes the attachment of L. rhamnosus to mucin and ECM proteins. The bacterial cell surface proteins that bind to ECM proteins have been termed MSCRAMMs (microbial surface cell recognition adhesion matrix molecule).Citation30) Many MSCRAMMs have been found in gram-positive pathogenic bacteria, including Cna from Staphylococcus aureus, and Enterococcus faecalis,Citation31,32) and Aaa from S. aureus.Citation33) Moreover, the interaction between these MSCRAMMs and ECM proteins exhibited submicromolar KD values. We propose that MBF is a member of the MSCRAMM family because MBF is cell-wall anchored protein and binds to several ECM proteins. In Lactobacillus spp., MBF might play an important role in the colonization in the intestinal lumen and the activation of certain probiotic effects, such as pathogen exclusion.Citation34,35) Because ECM components are thought to be available for interaction only after disruption of the epithelial barrier, such as during trauma, infection, or inflammation, the in vivo significance of the adhesion function is not yet clear.Citation3,36,37) Therefore, we intend to perform in vivo L. rhamnosus strain colonization studies to elucidate further the MBF-mediated adhesion processes and probiotic effects.
Supplemental material
The supplemental material for this paper is available at http://10.1080/09168451.2014.972325.
Supplemental Materials
Download Zip (952 KB)Acknowledgments
The authors would like to thank Kunihiko Taie for his technical assistance. The authors have no conflicts of interest to declare.
Additional information
Funding
Notes
Abbreviations: BSA, bovine serum albumin; DMEM, Dulbecco’s Modified Eagle’s Medium; ECM, extracellular matrix; FSMM, fermented Sumbawa mare milk; GI, gastrointestinal; His6-MBF, recombinant MBF fused to a 6×Histidine tag at the C terminus; ka, association rate; kd, dissociation rate; KD, dissociation constant; LB, Luria–Bertani; LGG, L. rhamnosus GG strain; MBF, mucus-binding factor; MRS, de Man–Rogosa–Sharpe; MSCRAMM, microbial surface components recognizing adhesive matrix molecule; MucBD, mucin-binding protein domain; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PBS-T, PBS-0.05% Tween 20; PCM, porcine colonic mucin; RUs, resonance units; SPR, surface plasmon resonance.
References
- Servin AL, Coconnier M-H. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract. Res. Clin. Gastroenterol. 2003;17:741–754.10.1016/S1521-6918(03)00052-0
- Vélez MP, De Keersmaecker SCJ, Vanderleyden J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 2007;276:140–148.10.1111/fml.2007.276.issue-2
- Van Tassell ML, Miller MJ. Lactobacillus adhesion to mucus. Nutrients. 2011;3:613–636.
- Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 2003;68:114–123.
- Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lähteinen T, Brouns SJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013;9:e1003683.10.1371/journal.pgen.1003683
- Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Adaptation factors of the probiotic Lactobacillus rhamnosus GG. Benef. Microbes. 2010;1:335–342.10.3920/BM2010.0032
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA. 2009;106:17193–17198.10.1073/pnas.0908876106
- von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, Tynkkynen S, Salminen S, de Vos WM, Palva A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 2010;76:2049–2057.10.1128/AEM.01958-09
- Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, de Vos WM, De Keersmaecker SC, Vanderleyden J. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012;78:185–193.10.1128/AEM.06192-11
- Vélez MP, Petrova MI, Lebeer S, Verhoeven TL, Claes I, Lambrichts I, Tynkkynen S, Vanderleyden J, De Keersmaecker SC. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol. Med. Microbiol. 2010;59:386–398.
- von Ossowski I, Satokari R, Reunanen J, Lebeer S, De Keersmaecker SC, Vanderleyden J, de Vos WM, Palva A. Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2011;77:4465–4472.10.1128/AEM.02497-10
- Bierne H, Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 2007;71:377–397.10.1128/MMBR.00039-06
- Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:238–245.10.1016/S0966-842X(02)02342-9
- Sabet C, Lecuit M, Cabanes D, Cossart P, Bierne H. LPXTG protein InlJ, a newly identified internalin involved in listeria monocytogenes virulence. Infect. Immun. 2005;73:6912–6922.10.1128/IAI.73.10.6912-6922.2005
- Lindén SK, Bierne H, Sabet C, Png CW, Florin TH, McGuckin MA, Cossart P. Listeria monocytogenes internalins bind to the human intestinal mucin MUC2. Arch. Microbiol. 2008;190:101–104.10.1007/s00203-008-0358-6
- Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology. 2006;152:273–280.10.1099/mic.0.28415-0
- MacKenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, Roos S, Walter J, Juge N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology. 2010;156:3368–3378.10.1099/mic.0.043265-0
- Etzold S, MacKenzie DA, Jeffers F, Walshaw J, Roos S, Hemmings AM, Juge N. Structural and molecular insights into novel surface-exposed mucus adhesins from Lactobacillus reuteri human strains. Mol. Microbiol. 2014;92:543–556.10.1111/mmi.2014.92.issue-3
- Etzold S, Kober OI, MacKenzie DA, Tailford LE, Gunning AP, Walshaw J, Hemmings AM, Juge N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 2014;16:888–903.10.1111/emi.2014.16.issue-3
- Shi T, Nishiyama K, Nakamata K, Aryantini NP, Mikumo D, Oda Y, Yamamoto Y, Mukai T, Sujaya IN, Urashima T, Fukuda K. Isolation of potential probiotic Lactobacillus rhamnosus strains from traditional fermented mare milk produced in Sumbawa Island of Indonesia. Biosci. Biotechnol. Biochem. 2012;76:1897–1903.10.1271/bbb.120385
- Kok J, van der Vossen JM, Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 1984;48:726–731.
- De Keersmaecker SC, Braeken K, Verhoeven TL. Perea Vélez M, Lebeer S, Vanderleyden J, Hols P. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 2006;72:4923–4930.10.1128/AEM.02605-05
- Biswas I, Gruss A, Ehrlich SD, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 1993;175:3628–3635.
- Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566.10.1016/0300-9084(88)90093-4
- Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 1989;55:3119–3123.
- Nishiyama K, Kawanabe K, Miyauchi H, Abe F, Tsubokawa D, Ishihara K, Yamamoto Y, Mukai T. Evaluation of bifidobacterial adhesion to acidic sugar chains of porcine colonic mucins. Biosci. Biotechnol. Biochem. 2014;78:1444–1451.
- Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, de Vos WM, Palva A, Lebeer S, Vanderleyden J, Dufrêne YF. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano. 2013;7:3685–3697.10.1021/nn400705u
- Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem. 2013;288:6849–6863.10.1074/jbc.M112.405530
- Toba FA, Visai L, Trivedi S, Lowy FD. The role of ionic interactions in the adherence of the Staphylococcus epidermidis adhesin SdrF to prosthetic material. FEMS Microbiol Lett. 2013;338:24–30.10.1111/fml.2012.338.issue-1
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229.
- Kang M, Ko YP, Liang X, Ross CL, Liu Q, Murray BE, Hook M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 2013;288:20520–20531.10.1074/jbc.M113.454462
- Ross CL, Liang X, Liu Q, Murray BE, Hook M, Ganesh VK. Targeted protein engineering provides insights into binding mechanism and affinities of bacterial collagen adhesins. J. Biol. Chem. 2012;287:34856–34865.10.1074/jbc.M112.371054
- Heilmann C, Hartleib J, Hussain MS, Peters G. The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 2005;73:4793–4802.10.1128/IAI.73.8.4793-4802.2005
- Horie M, Ishiyama A, Fujihira-Ueki Y, Sillanpää J, Korhonen TK, Toba T. Inhibition of the adherence of Escherichia coli strains to basement membrane by Lactobacillus crispatus expressing an S-layer. J. Appl. Microbiol. 2002;92:396–403
- Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 2007;9:356–367.10.1111/cmi.2007.9.issue-2
- Lorca G. Lactobacilli express cell surface proteins which mediate binding of immobilized collagen and fibronectin. FEMS Microbiol. Lett. 2002;206:31–37.10.1111/fml.2002.206.issue-1
- Styriak I, Nemcova R, Chang Y-H, Ljungh A. Binding of extracellular matrix molecules by probiotic bacteria. Lett. Appl. Microbiol. 2003;37:329–333.10.1046/j.1472-765X.2003.01402.x