Abstract
The bacterial community structure was compared between the third days’, one week’, and three weeks’ biofilm samples from the surface of a household toilet bowl. It was found that the PCR-DGGE band pattern of 16S rRNA gene was dramatically changed after the third day and was not further changed until three weeks. This result suggests that there are early and late colonizing bacterial groups. One of the early colonizers isolated from the third days’ sample was Rhizobium sp. R8, a closest relative to Rhizobium giardinii, which exhibited the highest biofilm formation activity in an artificial urine condition. R8 produced extracellular polysaccharides containing galactose, glucose, and mannose at the molar ratio of 8:1:1, which were probably responsible for the biofilm formation. Its excelled biofilm formation and urease activities together with the lack of nodulation and nitrogen fixing genes in R8 suggest that this strain has been specifically adapted to urine condition in a toilet bowl.
“Rhizobium latrinae” neither forms root nodules nor fixes atmospheric nitrogen, but instead exhibits high urease activity. This finding adds our knowledge about the ecological diversity of Rhizobia.

Biofilms are microbial community structures formed on solid surfaces, which are composed of cells and their extracellular matrices. Forming biofilms is considered an effective strategy for bacteria to proliferate in both favorable and hostile environments.Citation1,2) The biofilm formation, especially by multispecies microorganisms, is a complex phenomenon in which environmental conditions and microbial interactions govern its temporal and spatial variations. Multispecies interactions can influence the emergence and disappearance of species and therefore play an important role in the shaping of the biofilm communities.Citation3,4) For example, Pseudomonas aeruginosa rapidly spread over the surface via swarming and twitching motility and prevented Agrobacterium tumefaciens from invasion and adhesion.Citation5) Streptococcus mutans can serve as the initial colonizer on the tooth surface and together with an Actinomyces species promote biofilm growth of Lactobacillus in an oral multispecies biofilm.Citation6) Streptococcus gordonii is also a first colonizer on the tooth surface and provides conditions that allow later colonizers including Porphyromonas gingivalis, to adhere. Moreover, streptococcal cell wall polysaccharides have been shown to enable other oral bacteria to recognize and coagulate with streptococci, and ultimately form commensal dental plaque.Citation7,8) These multispecies biofilms are also ubiquitously formed in our living conditions.
Control and prevention of biofilm formation in public and household toilets are important from a hygienic point of view because they can cause biofouling, malodour, and staining and they represent a potential reservoir for pathogens. A great diversity of bacterial community in the biofilms formed on the surface of toilet bowls has been already demonstrated by both culture-dependent and culture-independent methods.Citation9–12) However, there is only limited information yet available for transient and temporal growth of biofilms on toilet bowls. The aim of this study was to shed light on the transition of biofilm formation on toilet bowls and characterize one of the early colonizers.
Materials and methods
Biofilm samples
Biofilm samples were collected from the inside surface of a household toilet according to the method as previously described.Citation10) Briefly, after careful cleaning the surface of the toilet bowl, the toilet was normally used by a family for two weeks. The inside surface of toilet bowl was carefully swabbed by a sterile cotton fabric (Japan Textile Evaluation Technology Council) for sampling. Samples were taken on the third day and after one and three weeks. Biofilms were released from the cotton fabric by vigorous vortexing in an aliquot of LP diluent (Nihon Pharmaceutical, Co., Ltd) and spread on R2A agar plates after appropriate dilutions. Bacterial colonies were grown on the plate for a week at 30 °C.
Culture media and bacterial strains
R2A medium was used for isolation of bacteria and maintenance culture. R2A contains per liter 0.5 g each of Bacto proteose peptone No.3 (Difco), Bacto yeast extract (Difco), casamino acid, glucose, soluble starch, and 0.3 g each of K2HPO4, sodium pyruvate, and 0.05 g of MgSO4–7H2O (pH 7.0–7.4). Artificial urine (AU) medium mimicked human urine condition.Citation13) AU medium contained per liter 1 g of Bacto peptone, 5 g of Bacto yeast extract, 2.1 g of NaHCO3, 10 g of urea, 70 mg of uric acid, 0.8 g of creatinine, 5.2 g of NaCl, 2.3 g of Na2SO4, 0.95 g of KH2PO4, 1.2 g of K2HPO4, 1.3 g of NH4Cl, 0.37 g of CaCl2·2H2O, 0.49 g of MgSO4·7H2O, 1.2 mg of FeSO4·7H2O, 110 μL of lactic acid, 2 mL of 6 N HCl, and pH was adjusted to 6.5. TSB and TY media were used for nutrient rich culture. TSB contained per liter of 17 g pancreatic digest of casein, 3 g of enzymatic digest of soybean meal, 5 g of NaCl, 2.5 g each of K2HPO4 and dextrose. pH was adjusted to 7.3. TY contained per liter 5 g of Bacto tryptone (Difco), 3 g of Bacto yeast extract, and 0.83 g of CaCl2–2H2O (pH 6.8–7.0).
Rhizobium giardinii. NBRC 107135 [=H152T]Citation14) was used as a reference strain. Rhizobium strains H152T and R8 (waiting for NBRC strain number) were usually cultured in TY medium at 30 °C. When exopolysaccharides were prepared, YM medium was used. YM medium contained per liter 10 g mannitol, 4 g CaCO3, 0.4 g Bacto yeast extract, 0.5 g KH2PO4, 0.2 g MgSO4–7H2O, 0.1 g NaCl (pH 6.8–7.0). YMA contained per liter 15 g of agar in YM for preparation of solid medium. Other isolated strains were cultured in R2A medium. Escherichia coli DH5α was used as a host strain for general DNA cloning. A cloning vector pUC19 was used in E. coli DH5α.
PCR-DGGE analysis
DNA extraction from the biofilm samples was carried out using ISOIL for Beads Beating (Nippon Gene). Extracted DNA was used as templates for PCR targeting a part of 16S rRNA gene. The nucleotide sequences of the primers were as follows: forward primer with GC-clump (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3′) and reverse primer 5′-ATTACCGCGGCTGCTGG-3′. The reaction mixture was composed of 5 μL of 10xPCR buffer, 5 μL of 2 mM dNTP, 3 μL of 25 mM MgSO4, 5 ng of template DNA, 1.5 μL each of forward and reverse primers, 1 μL of KOD Plus-Neo DNA polymerase (Toyobo), and sterile water up to 50 μL in total volume. The template DNA was first denatured for 2 min at 94 °C, followed by 20 cycles of the following steps: denaturation for 10 s at 98 °C, annealing for 30 s from 65 to 55 °C (touchdown by −0.5 °C cycle−1), and extension for 12 s at 68 °C (MJ MiniTM, Bio-Rad). The 20 cycles were further followed by 10 cycles of above temperature programs with a constant annealing temperature 55 °C. Denaturant gradient gel electrophoresis, DGGE, was performed using D-CodeTM universal mutation detection system (Bio-Rad). Denaturant gradient was formed from 30 to 60% where 100% corresponded to 7 M urea and 40% formamide in an 8% polyacrylamide gel (16 cm × 16 cm × 1 mm thickness in 0.5 × TAE). Electrophoresis was carried out at 200 V for 4 h at 58 °C in a tank containing 7 L of 0.5 × TAE buffer. After electrophoresis, the gel was stained for 30 min with 0.01% (v/v) SYBR Gold solution (Molecular Probes), rinsed with water, and photographed on a Fluoroimager 595 (molecular dynamics).
16S rRNA gene sequence analysis of the bacterial isolates
Genomic DNA was prepared by InstageneTM MATRIX (Bio-Rad) and used for PCR amplification of 16S rRNA genes. The reaction mixture was composed of 5 μL of 10 × PCR buffer, 5 μL of 2 mM dNTP, 2 μL of 25 mM MgSO4, 2 μL of template DNA, 1 μL [10 pmol/μL] each of 16S rRNA gene general forward 27F and reverse 1492R primers, 1 μL of KOD Plus-Neo DNA polymerase (Toyobo), and sterile water up to 50 μL in total volume. The templates were first denatured for 2 min at 94 °C, followed by 29 cycles of the following steps: denaturation for 10 s at 98 °C, annealing for 30 s at 52 °C, and extension for 45 s at 68 °C. The 29 cycles were followed by a final 3 min extension at 68 °C. Approximately 1,500-bp PCR products were purified with a QIAquick gel extraction kit (Qiagen) and cloned into the SmaI gap of pUC19. The nucleotide sequence of the 16S rRNA gene was determined by BigDye terminator cycle sequencing kit ver. 3 on an ABI 3130 DNA sequencer (Applied biosystems). The sequence data were analyzed by GENETYX-WIN version 5.1.1 (Toyobo) and nucleotide BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The 16S rRNA gene sequences of relevant 54 Rhizobia were retrieved from the NCBI database to construct a phylogenetic tree. Phylogenetic trees based on multiple sequence alignments were generated by EBI Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and neighbor-joining method using MEGA version 5.5 with 1,000 bootstrap trials.
Biofilm formation assay
Pre-culture of each strain was inoculated at 1% into 200 μL of R2A medium in polystyrene 96-well microplates (#167008, Thermo Scientific™ Nunc™) or 3 mL in porcelain crucibles and statically grown to form biofilms at 30 °C for 24 and 48 h. Planktonic cells in the culture liquid were quantified by measuring optical density at 595 nm (OD595), and then, the culture was removed by pipet, and the biofilms formed on surface of the vessels were washed with distilled water. The biofilms were stained by 0.1% (w/v) crystal violet for 10 min and washed twice with distilled water. Adherent crystal violet was extracted by an equal volume of 33% acetic acid for 10 min. Biofilm formation activity was determined by measuring absorbance of the solution at 595 nm (A595) with a microplate reader (Sunrise, TECAN).
Urease assay
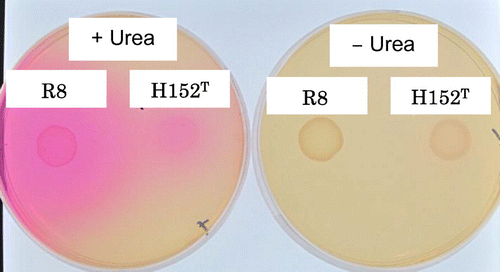
Bacterial cells of overnight culture grown in TSB medium were washed twice with equal amounts of 1 × phosphate-buffered saline (150 mM NaCl, 10 mM Na2HPO4, 20 mM NaH2PO4, pH 7.0), and an aliquot of the sample (5.0 μL) was spotted on a 1.5% Christensen urea agar plate containing, per liter 1 g of Bacto peptone, 1 g of glucose, 0.5 g of NaCl, 1.2 g of Na2HPO4, 0.8 g of NaH2PO4, 12 mg of phenol red, and 20 g of urea. A plate without 20 g urea was prepared for negative control experiment. After two days’ cultivation at 30 °C, urease activity was detected as the formation of a red-colored clear zone around the colony.Citation13)
PCR examination of nodC and nifH gene
Existence of nodC and nifH genes was examined by PCR using NEB One TaqTM 2X master mix. The nucleotide sequences of the primers were as follows: nodCf (5′-AYGTHGTYGAYGACGGTTC-3′) and nodCr (5′- CGYGACAGCCANTCKCTA TTG-3′); nifHf (5′-TACGGNAARGGSGGNATCGGCAA-3′) and nifHr (5′-AGCATGTCYTCSAGYTCNTCCA-3′).Citation15) DNA templates were first denatured for 2 min at 94 °C, followed by 30 cycles of the following steps: denaturation for 10 s at 94 °C, annealing for 30 s at 55 °C, and extension for 1 min at 68 °C. The 30 cycles were followed by a final 5 min for extension at 68 °C.
Nodule formation assay
Seeds of Phaseolus vulgaris “Koi-midori” (Takii & Co., Ltd) were surface-sterilized by soaking for 10 min in 2% (w/v) sodium hypochlorite and 0.02% Tween 20 solution, followed by five times wash with sterile distilled water and germinated on B&D [nitrogen free] medium containing 0.7% agar.Citation16) After germination, primary roots of 2-day-old seedlings were inoculated by dripping 200 μL bacterial suspension at about 108 colony forming units/mL. The plants were transferred to 400 mL plant boxes containing expanded vermiculite with an appropriate amount of B&D medium. The plants were cultured for six weeks in a growth chamber at 25 °C with a 12-h photoperiod.Citation17) The experiments were performed in triplicates. R. giardinii H152T was used as a positive control for nodulation, and water treatment was used as a negative control.
Scanning electron microscopy
Planktonic cell suspension was spotted and fixed on a poly-L-Lysine-coated cover glass. Biofilms were grown in the 35-mm-diameter glass bottom dish (AGC Techno Glass) containing R2A medium at 30 °C for 48 h. Both samples were fixed with 2% glutaraldehyde and dehydrated with ethanol by gradually increasing the concentration, 50, 70, 80, 90, 95, and 100% followed by isoamyl acetate. After CO2 critical point drying with JCPD-5 (JEOL), samples were sputter-coated with gold (JFC-1100, JEOL). Scanning electron micrographs were obtained with S-2400 (Hitachi).
Physiological characterization of Rhizobium strains
Physiological characterization of R8 was performed according to the standard methodCitation14,18) with several modifications. Most of the biological tests were done using ϕ 18-mm test tubes with 3 mL basal medium containing per liter 1 g of KH2PO4, 1 g of K2HPO4, 0.01 g of FeCl3·6H2O, 0.2 g of MgSO4·7H2O, 1 g of (NH4)2SO4, 0.1 g of CaCl2, and one each of carbon sources at 0.1% (w/v). When amino acids were tested for nitrogen sources, ammonium sulfate was eliminated and mannitol was used as a carbon source. Rhizobium sp. R8 or R. giardinii H152T pre-culture in TY medium was washed by a newly prepared above test medium and inoculated at initial absorbance at 595 nm, OD595 = 0.01. The test tube cultures were shaken at 120 rpm, 28 °C for 4 days. Utilization of carbohydrate and organic acid were determined by cell growth measuring OD595. Plate tests for antibiotics resistance and NaCl tolerance, pH dependence, and requirement for calcium were conducted on TY-agar plate.
Preparation and analysis of membrane proteins
Biofilm-associated cells were prepared from inner surface of flasks after standing culture in R2A medium at 30 °C for 48 h. Planktonic cells were prepared by shaking culture in TY medium because R8 formed aggregates even in R2A shaking culture. The inoculum size was 1% from fully grown pre-culture. After spin down by centrifuge, cells were re-suspended in 50 mM Tris-HCl (pH 6.8) and disrupted by Multi-beads shocker (Yasui Kikai) using 0.1-mm glass beads. Disruption condition was twelve cycles of 60 s on and 60 s off at 2,500 rpm. After settle down and removal of glass beads by gravity, the disrupted cell suspension was centrifuged at 11,000 × g for 5 min to eliminate intact cells. Cytoplasmic and membrane fractions were separated by ultracentrifuge (4 °C, 604,000 × g, 120 min). Each fraction was boiled in SDS sample buffer (50 mM Tris-HCl pH 6.8, 6% 2-mercaptoethanol, 10% glycerol, 2% SDS, and 0.01% bromophenol blue) and analyzed by SDS-PAGE using 12% polyacrylamide gel. Electrophoresis was carried out at 100 V for 2.5 h. Protein bands were visualized by Coomassie Brilliant Blue R-250 staining. Proteins from major bands were extracted overnight in 1% SDS/20 mM Tris-Cl (pH 8.0) and applied for N-terminal amino acid sequence analysis by Procise 492 (PerkinElmer) or Procise 491 cLC (Applied Biosystems). The protein sequence was analyzed by protein BLAST homology search program, http://blast.ncbi.nlm.nih.gov/Blast.cgi.
Preparation and analysis of EPS
Strain R8 was grown at 28 °C for 96 h, in yeast extracts mannitol agar (YMA) plate,Citation19) the colonies were scraped off with disposable cell scraper (#99002, TPP Techno Plastic Products) and suspended in 40 mL distilled water. Bacterial cells were removed by centrifugation and further filtration (0.22 μm pore size Millex GV, Millipore), and clear supernatant containing extracellular polysaccharide (R8-EPS) was pooled. R8-EPS was precipitated by addition of equal volume of ethanol and stored overnight at 4 °C. Resulting precipitates were recovered by centrifuge (7,850× g, 4 °C, 20 min) and lyophilized. Purity of polysaccharides was spectrometrically verified by no apparent absorbance at 254 and 280 nm. The recovery yield was about 10 mg/two plates. R8-EPS was hydrolyzed by 2 M trifluoroacetic acid for 4 h at 100 °C under nitrogen atmosphere. Pyridylamination of hydrolyzed R8-EPS was performed according to the method of Hase et al.Citation20) with slight modifications. The pyridylamino (PA)-derivative sugars were separated using a reverse phase column (4.6 × 250 mm CAPCELL PAK MG, Shiseido) with 20 mM ammonium acetate buffer, pH 7.5, containing 2.0% acetonitrile, at a flow rate of 1.0 mL/min, 40 °C. For the detection of PA-derivative sugar, excitation and emission wavelength of 310 nm and 380 nm was used, respectively.
Sequence data submission
The 1,467 nucleotides sequence data of R8 16S rRNA gene have been registered to DDBJ/EMBL/GenBank under accession number LC005488.
Accession numbers of the 16S rRNA sequence data obtained from DDBJ/EMBL/GenBank
ABB19816, KJ801857, JQ689178, JQ660243, KJ631291, EU730917, JF913979, KF973257, KJ631291, KF358264, EF035074, U71078, U89832, EF061096, AY738130, D14501, AY626395, FJ839677, U28916, U29386, EF141340, DQ835306, AY509899, AM931436, DQ100063, Y10170, AF364068, U89817, AF003375, AF364069, U86343, EU867317, EU256404, EU256434, AM181745, AF041447, D13431, U07934, D14514, X67231, D14509, GU565534, U86344, Y17047, U45329, AY341343, EF440185, EU781656, X73041, AB247615, FJ969841, Z30542, EF125187, DQ454123, EU056823, EF363715, DQ855276, AF025852, EU074168, D11343, GU128881, U69638, X87273.
Results and discussion
Change in bacterial community structure of the biofilm samples
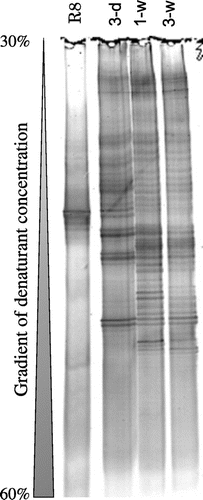
Culture-independent analysis of bacterial community structure in the biofilms formed on toilet bowls was performed by PCR-DGGE using short-length 16S rRNA gene fragments (approximately 200 bp) (Fig. ). It was shown that the DNA band patterns are quite similar between one and three weeks’ biofilm samples (1-w and 3-w), suggesting the stability of microbial community structure during this time period. On the other hand, only a part of them were observed in the third days’ sample (3-d). These results suggest that there are a number of later colonizing bacterial groups that prefer conditions formed by early colonizing bacterial groups in 3-d sample. Analysis of these early coming bacterial groups should provide us with knowledge about the initial process of forming biofilms on toilet bowls and also might be a key to control and eliminating stable and recalcitrant biofilm formation. We were thus prompted to isolate early biofilm forming bacteria from the 3-d sample.
Isolation of early colonizing bacteria
Eleven candidate strains, R1-R11, were isolated in R2A at 30 °C from the 3-d biofilm sample. The nucleotide BLAST analysis of each partial 16S rRNA gene sequence revealed that these are, R1, Chryseobacterium sp. (99.4%: 1371/1378 bp identical to Chryseobacterium sp. L7-15, ABB19816); R2, Microbacterium sp. (99.2%: 1340/1351 bp identical to Microbacterium sp. N54, KJ801857); R3, Microbacterium sp. (97.6%: 1335/1367 bp identical to Microbacterium trichothecenolyticum A3RC1, JQ689178); R4, Sphingomonas sp. (98.5%: 1324/1344 identical to Sphingomonas sp. S7-655, JQ660243); R5 Microbacterium sp. (98.8% 1348/1365 bp identical to M. trichothecenolyticum N3G-5, KJ631291); R6, Sphingomonas sp. (99.4% 1333/1341 identical to Sphingomonas yunnanensis 215, EU730917); R7, Sphingomonas sp. (99.5% 133/1340 bp identical to S. yunnanensis 215, EU730917); R8, Rhizobium sp. (95.5% 1300/1360 bp identical to R. giardinii D30, JF913979); R9, Microbacterium sp. (95.1% 770/809 identical to Microbacterium foliorum XLST-6, KF973257); R10, Microbacterium sp. (97.6% 774/793 bp identical to M. trichothecenolyticum N3G-5, KJ631291); and R11, Microbacterium sp. (93.9% 1320/1405 bp identical to Microbacterium oxydans M2-2, KF358264), respectively. Taking ambiguity of several nucleotide sequences into consideration and similar biofilm formation activities shown below, it is probable that strains R3 and R5, and R6 and R7 are the same strain, respectively.
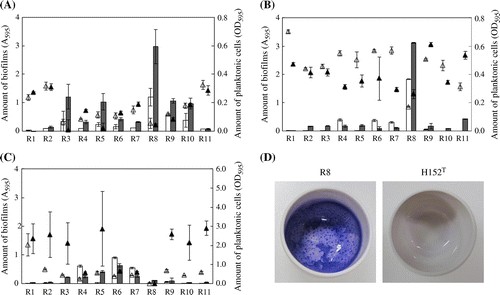
Biofilm forming activity was tested for the eleven strains in three different media, AU, R2A, and TSB (Fig. (A)–(C)). It was found that Rhizobium sp. R8 exhibited the highest biofilm forming activity in both AU and R2A media, while did not grow on nutrient rich TSB medium. Microbacterium sp. R3, R5, R9, and R10 showed the second best activity in AU medium. Sphingomonas sp. R4, R6, and R7 had marginal biofilm forming activity in all media. Chriseobacterium sp. R1 and Microbacterium sp. R2 were poor biofilm former in all the three medium conditions. All the strains showed vigorous growth in TSB medium; however, the biofilm formation was generally poor except for strains R4, R6, and R7. R8 also formed robust biofilms on porcelain crucible whose surface is similar to the toilet bowl (Fig. (D)). We thus chose R8 for further characterization and identification of its cellular factors related to the biofilm formation on abiotic solid surfaces. Comparison of the PCR-DGGE band pattern of R8 with 3-d biofilm sample suggested that R8 was not the most dominant bacterium (Fig. ). We have often faced to the limitation of cultivation method for environmental bacteria, such as the most dominant bacteria could be VBNC, viable but non-culturable.
Fig. 2. Biofilm forming activity of isolated bacteria.
Notes: Three medium conditions were examined: (A) AU; (B) R2A; (C) TSB. Bar graphs (A595) show the amount of biofilms formed in polystyrene 96-well microplate. Error bar shows SD in triplicate experiments. Triangle shows amount of planktonic cells (OD595). Open symbol; 24 h, closed symbol: 48 h. (D) biofilm formation by Rhizobium strains on porcelain crucibles. R8 and H152T are isolated Rhizobium strain and the type strain of R. giardinii, respectively. Each strain was cultured in 3 mL R2A medium at 30 °C for 24 h. Biofilms formed on the surface were visualized by 0.1% crystal violet staining for 10 min.
Phylogenetic tree analysis and physiological characterization of Rhizobium sp. R8
In order to identify the strain R8, we first determined almost full sequence of the 16S rRNA gene (1,467 bp) and constructed a phylogenetic tree with other related Rhizobium type strains (Fig. ). The 16S rRNA gene sequence of R8 had the highest similarities 99.3% (1,146/1,154 bp) to that of R. giardinii H152T, followed by 99.2% (1,143/1,152 bp) to Rhizobium herbae CCBAU 83011T. We next compared physiological characterization of R8 with R. giardinii H152T. R. giardinii H152T was isolated from a nodule of a leguminous plant Phaseolus bulgaris grown in Bretennieres, France.Citation14) R8 exhibited almost similar assimilation ability of carbohydrates and amino acids to those of H152T but was slightly different from H152T in regard to the growth ability on D-dulcitol, requirement of calcium, and drug resistance for kanamycin, gentamycin, and erythromycin (Table ). Moreover, growth inability on citrate and Luria-Bertani medium and sensitivity to 2% NaCl were different feature from Rhizobium herbae CCBAU 83011T.Citation18) The most significant trait of R8 that differed from R. giardinii was urease activity (Fig. ). Tremendous urease activity should benefit R8 to dominate in the urine conditions on a toilet bowl. It may be also noteworthy that R8 cells formed abundant aggregates when cultured in shaking flask containing R2A medium, while H152T grew in uniformly dispersed planktonic state.
Fig. 3. A phylogenetic tree by neighbor-joining method showing the relationships of genus Rhizobium based on 16S rRNA gene sequences.
Note: Bootstrap probabilities (shown as percentages at junctions) were determined from 1,000 samplings.
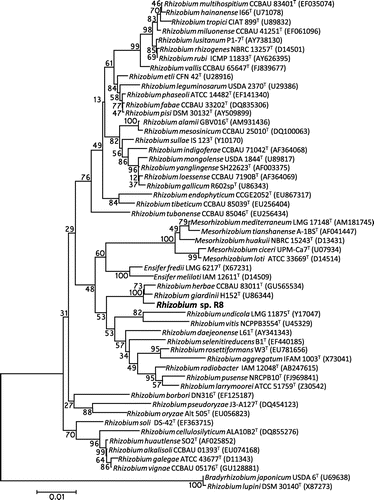
Table 1. Discriminative features of strain R8 and related strains in the genus Rhizobium.
Nodule forming activity
There is increasing number of reports for Rhizobium strains that are isolated not from plant roots but from various environments such as activated sludge and bioreactor or contaminated soil environments with aniline, petroleum oil, and triazophos. In such cases, these Rhizobium strains often lack nod and/or nif genes responsible for forming nodules and fixing nitrogen on the roots of leguminous plants.Citation21−23) R8 was isolated from a household toilet bowl which was different from plant-habiting area. We asked if R8 also did not harbor nodC and nifH genes. nodC encodes N-acetylglucosaminyltransferase that is one of the key enzymes to produce Nod factors. nifH encodes a dinitrogenase reductase subunit responsible for nitrogen fixation. nodC and nifH are the most widely used nodulation and nitrogen fixation gene markers, respectively.Citation15) It was found that PCR for amplifying nodC was successful when using template genomic DNA prepared from H152T but not successful from R8 (figures not shown). As to nifH, no positive PCR product was detectable for both H152T and R8 strains.Citation15) These results suggest that R8 had lost nodC and nifH genes. Furthermore, nodule forming activity was examined for R. giardinii strains H152T and R8 by inoculating them to the seedlings of a leguminous plant P. vulgaris. It was clearly shown that H152T but not R8 had ability to form nodules on the roots (Fig. ).
Fig. 5. Nodule forming activity.
Notes: Nodules were observed on the root of P. vulgaris inoculated by H152T but not by R8. Arrows indicate nodules. Size bar indicates 2 cm. Experiments were performed in triplicates.

Rhizobium strains can usually establish a symbiotic interaction with leguminous plants under nitrogen-limited soil conditions.Citation24,25) This host plant-specific interaction results in development of root nodules in which the Rhizobium strains differentiate into nitrogen fixing bacteroids.Citation26) It should be rational decision for Rhizobium sp. R8 to have lost abilities of energy consuming nodule formation and nitrogen fixation and acquired high urease activities in urine (soluble nitrogen source) rich conditions. On the other hand, popular nodule forming Rhizobium strains including, R. giardinii H152T, may maintain urease activity to be ready for the occasion of plant non-associated growth in the environments.
Membrane protein analysis of R8
There are examples of comparative proteomic analyses of biofilm-associated cells and planktonic cells. Production of outer membrane proteins, including OmpA-related protein, OmpW family protein, TonB-dependent receptor proteins, and non-fimbrial adhesion YapH, was clearly increased in biofilm-associated cells.Citation27) Proteins involved in the motility complex, including the flagellins (FlaA, FlaB), the filament cap (FliD), the basal body (FlgG, FlgG2), and the chemotactic protein (CheA), all exhibited higher levels of expression in biofilms than found in stationary-phase planktonic cells of Campylobacter jejuni.Citation28) We hypothesized that the production level of specific proteins including proteinaceous adhesion factors should be increased in membrane or cell surfaces upon formation of biofilms by R8.
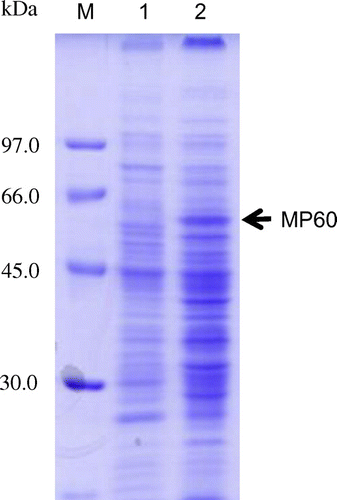
Membrane fractions of biofilm-associated cells and planktonic cells were prepared and analyzed their protein production profiles by SDS-PAGE (Fig. ). There were several membrane proteins produced in different amounts between the samples. It was found that a protein band at approximately 60 kDa, namely MP60, was the most specifically overproduced in the biofilm-associated cells. The N-terminal amino acid sequence of MP60 was determined as 1-KVFGRIELAA, and this sequence was analyzed by protein BLAST program. Although there were proteins whose amino acid sequence was partially similar to this sequence, none of them shared similarity at the N-terminal position. A hypothetical protein (WP_017996705; 143 a.a. = 15.2 kDa) from Rhizobium leguminosarum had the sequence 31-KVFARIDFAA; however, this protein seemed to be neither a membrane nor a secretion protein based on the analysis by signal peptide prediction program SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). It is probable that MP60 is a unique membrane protein of R8.
Exopolysaccharides produced by R8
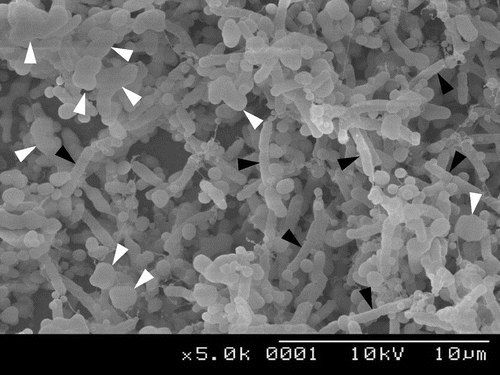
When we observed the cell structure of R8 by scanning electron microscope, we noticed that R8 cells were incased in abundant extracellular polymers (Fig. ). It was known that Rhizobacterium bacteria effectively produced exopolysaccharides in culture media containing mannitol.Citation29) When R8 was grown on YMA plate, the colonies became slimy and covered by significant amount of jelly-like exopolysaccharides. The amount of biofilms formed by R8 was also increased by twofolds (A595 = 5.3) upon addition of 1% mannitol to R2A medium, suggesting that these mannitol-dependent exopolysaccharides, namely EPS-R8, are also functional biofilm matrices. This dramatic change in the colony morphology and increase in the biofilm formation were not observed for H152T. We were thus prompted to prepare and analyze the structure of EPS-R8.
Fig. 7. SEM images of Rhizobium sp. R8 biofilm-associated cells.
Notes: White arrows indicate cells that probably produce large amount of EPS, while black arrows indicate cells that produce small amount of EPS.
About 10 mg (dry weight) EPS-R8 was prepared from slime colonies formed on two YMA plates. HPLC analysis of 2-aminopyridyne sugars showed that EPS-R8 is composed of galactose, glucose, and mannose at the ratio of 8:1:1. There is a report that R. leguminosarum, Rhizobium meliloti, Rhizobium phaseoli, and Rhizobium trifolii commonly produced gel-forming capsular polysaccharides (CPS), which contained galactose, glucose, and mannose at a constant ratio of 4:1:1.Citation29,30) This ratio was reported unchangeable under different concentration of mannitol. These CPS were reported insoluble in water and prepared from cell surfaces by 1 N NaOH treatment. On the other hand, the EPS-R8 was soluble in water and recovered from cell-free supernatants by simple centrifuge of cell suspension. The CPS structure was shown pseudo-double helix with hexasaccharide repeating unit composed of a main chain →4)-Glc-(1→3)-Man-(1→3)-Gal-(1→, which is doubly branched by single Gal-(1→ side chain at position O-2 and by a Gal-(1→4)-Gal-(1→ side chain at position O-6 of the glucose residue.Citation31,30) EPS-R8 may contain longer or more galactosyl side chains, which enables to be more soluble in water than CPS.
R. meliloti also produced acidic EPS composed of D-glucose, D-galactose, pyruvic acid, and succinic acid in the molar ratios 7:1:1:1 which is called succinoglycan.Citation32) On the other hand, R. leguminosarum, R. trifolii, and R. phaseoli produced another group of acidic EPS composed of D-glucose, D-galactose, D-glucuronic acid, and pyruvic acid in the molar ratios 5:1:2:2.Citation33) Production of these high molecular weight EPS, together with low molecular weight β-l, 2-glucans and acidic oligosaccharides, has been shown to be important for nodule formation by Rhizobium bacteria.Citation34) Our experimental data demonstrated that EPS-R8 did not contain succinate, pyruvate, and uronic acids. This result is not inconsistent with the fact that R8 had lost genes essential for nodulation and nitrogen fixation. It remains to be elucidated whether MP60 together with surface EPS-R8 plays crucial roles staying against water flush and recruiting some friendly neighbors in the multispecies toilet bowl biofilms.
Proposal of “Rhizobium latrinae” sp. nov.
The 16S rRNA gene of Rhizobium strain R8 shares significantly high sequence similarities with R. giardinii (99.3%) and R. herbae (99.2%), suggesting R8 is a member of species giardinii or herbae. However, its robust biofilm formation activity, high urease activity, production of novel exopolysaccharide, and loss of nodule forming activity and nifH and nodC genes are specific traits of R8. Although DNA–DNA hybridization experiment remains to be performed, in order to avoid confusion of R8 with above two species, we propose a non-validated novel species name “Rhizobium latrinae” R8 sp. nov. The word “latrina” means “toilet”.
Acknowledgment
The authors are thankful to Mr Yasunori Takahashi for technical assistance.
References
- Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004;236:163–173.10.1111/fml.2004.236.issue-2
- Shimada K, Itoh Y, Washio K, Morikawa M. Efficacy of forming biofilms by naphthalene degrading Pseudomonas stutzeri T102 toward bioremediation technology and its molecular mechanisms. Chemosphere. 2012;87:226–233.10.1016/j.chemosphere.2011.12.078
- Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 2012;36:990–1004.
- Rendueles O, Ghigo JM. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012;36:972–989.
- An D, Danhorn T, Fuqua C, Parsek MR. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Nat. Acad. Sci. USA. 2006;103:3828–3833.10.1073/pnas.0511323103
- Filoche SK, Anderson SA, Sissons CH. Biofilm growth of Lactobacillus species is promoted by Actinomyces species and Streptococcus mutans. Oral Microbiol. Immunol. 2004;19:322–326.10.1111/omi.2004.19.issue-5
- Cisar JO, Sandberg AL, Reddy GP, Abeygunawardana C, Bush CA. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral Actinomyces and streptococcal lectins. Infect. Immun. 1997;65:5035–5041.
- Xu DQ, Thompson J, Cisar JO. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J. Bacteriol. 2003;185:5419–5430.10.1128/JB.185.18.5419-5430.2003
- Egert M, Schmidt I, Bussey K, Breves R. A glimpse under the rim – the composition of microbial biofilm communities in domestic toilets. J. Appl. Microbiol. 2010;108:1167–1174.10.1111/jam.2010.108.issue-4
- Mori M, Gomi M, Matsumune N, Niizeki K, Sakagami Y. Biofilm-forming activity of bacteria isolated from toilet bowl biofilms and the bactericidal activity of disinfectants against the isolates. Biocontrol Sci. 2013;18:129–135.10.4265/bio.18.129
- Ojima M, Toshima Y, Koya E, Ara K, Tokuda H, Kawai S, Kasuga F, Ueda N. Hygiene measures considering actual distributions of microorganisms in Japanese households. J. Appl. Microbiol. 2002;93:800–809.10.1046/j.1365-2672.2002.01746.x
- Pitts B, Stewart PS, McFeters GA, Hamilton MA, Willse A, Zelver N. Bacterial characterization of toilet bowl biofilm. Biofouling. 1998;13:19–30.10.1080/08927019809378368
- Oki K, Washio K, Matsui D, Kato S, Hirata Y, Morikawa M. The role of urease activity on biofilm formation by Staphylococcus sp. T-02 isolated from the toilet bowl. Biosci. Biotechnol. Biochem. 2010;74:583–589.10.1271/bbb.90796
- Amarger N, Macheret V, Laguerre G. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int. J. Syst. Bacteriol. 1997;47:996–1006.10.1099/00207713-47-4-996
- Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2001;147:981–993.
- Broughton WJ, Dilworth MJ. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971;125:1075–1080.
- Yoon JH, Kang SJ, Yi HS, Oh TK, Ryu CM. Rhizobium soli sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2010;60:1387–1393.10.1099/ijs.0.013094-0
- Ren D, Wang ET, Chen WF, Sui XH, Zhang XX, Liu HC, Chen WX. Rhizobium herbae sp. nov. and Rhizobium giardinii-related bacteria, minor microsymbionts of various wild legumes in China. Int. J. Syst. Evol. Microbiol. 2011;61:1912–1920.10.1099/ijs.0.024943-0
- Breedveld MW, Zevenhuizen LP, Canter Cremers HC, Zehnder AJ. Influence of growth conditions on production of capsular and extracellular polysaccharides by Rhizobium leguminosarum. Antonie van Leeuwenhoek. 1993;64:1–8.10.1007/BF00870915
- Hase S, Ikenaka T, Matsushima Y. Structure analyses of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem. Biophys. Res. Commun. 1978;85:257–263.10.1016/S0006-291X(78)80037-0
- Gu T, Sun LN, Zhang J, Sui XH, Li SP. Rhizobium flavum sp. nov., a triazophos-degrading bacterium isolated from soil under the long-term application of triazophos. Int. J. Syst. Evol. Microbiol. 2014;64:2017–2022.10.1099/ijs.0.061523-0
- Kaur J, Verma M, Lal R. Rhizobium rosettiformans sp. nov., isolated from a hexachlorocyclohexane dump site, and reclassification of Blastobacter aggregatus Hirsch and Müller 1986 as Rhizobium aggregatum comb. nov. Int. J. Syst. Evol. Microbiol. 2011;61:1218–1225.10.1099/ijs.0.017491-0
- Zhang X, Li B, Wang H, Sui X, Ma X, Hong Q, Jiang R. Rhizobium petrolearium sp. nov., isolated from oil contaminated soil. Int. J. Syst. Evol. Microbiol. 2012;62:1871–1876.10.1099/ijs.0.026880-0
- Gibson AH, Pagan JD. Nitrate effects on the nodulation of legumes inoculated with nitrate-reductase-deficient mutants of Rhizobium. Planta. 1977;134:17–22.10.1007/BF00390088
- Streeter JG. Nitrate inhibition of legume nodule growth and activity: I. Long term studies with a continuous supply of nitrate. Plant Physiol. 1985;77:321–324.10.1104/pp.77.2.321
- Smit G, Swart S, Lugtenberg BJ, Kijne JW. Molecular mechanisms of attachment of Rhizobium bacteria to plant roots. Mol. Microbiol. 1992;6:2897–2903.10.1111/mmi.1992.6.issue-20
- Zimaro T, Thomas L, Marondedze C, Garavaglia1 BS, Gehring C, Ottado J, Gottig N. Insights into Xanthomonas axonopodis pv. citri biofilm through proteomics. BMC Microbiol. 2013;13:186.10.1186/1471-2180-13-186
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM. Proteomic analysis of campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 2006;188:4312–4320.10.1128/JB.01975-05
- Zevenhuizen LPTM. Gel-forming capsular polysaccharide of fast-growing rhizobia: occurrence and theological properties. Appl. Microbiol. Biotechnol. 1984;20:393–399.
- Zevenhuizen LPTM, van Neerven ARW. (1→2)-β-D-glucan and acidic oligosaccharides produced by Rhizobium meliloti. Carbohydr. Res. 1983;118:127–134.10.1016/0008-6215(83)88041-0
- Lee EJ, Chandrasekaran R. The, “pseudo double-helical” structure of the gel-forming capsular polysaccharide from Rhizobium trifolii. Carbohydr. Res. 1992;231:171–183.10.1016/0008-6215(92)84017-M
- Åman P, McNeil M, Franzén L, Darvill AG, Albersheim P. Structural elucidation, using h.p.l.c.-m.s. and g.l.c.-m.s., of the acidic polysaccharide secreted by Rhizobium meliloti strain 1021. Carbohydr. Res. 1981;95:263–282.10.1016/S0008-6215(00)85582-2
- Sutherland IW. Chapt 16 microbial exopolysaccharides. In: Dumitriu S, editor. Polysaccharides. 2nd ed. New York (NY): Marcel Dekker; 2005. p. 431–454.
- González JE, York GM, Walker GC. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146.10.1016/S0378-1119(96)00322-8