Abstract
Pseudomonas sp. 61-3 accumulates two types of polyhydroxyalkanoates (PHAs), poly(3-hydroxybutyrate) [P(3HB)], and poly(3HB-co-3-hydroxyalkanoates) [P(3HB-co-3HA)], and some proteins associated with their PHA granules have been identified. To date, PhaFPs (GA36) and PhaIPs (GA18) were identified from P(3HB-co-3HA) granules. In this study, the gene encoding GA24 associated with P(3HB) granule was identified as phbPPs. PhbPPs was composed of 192 amino acids with a calculated molecular mass of 20.4 kDa and was assumed to be a phasin. phbFPs gene and unknown ORF were also found on phb locus. PhbFPs was anticipated to be the transcriptional repressor of phbPPs gene. PhbPPs was bound to the P(3HB-co-3HA) granules with 3HB composition of more than 87 mol%, and PhaIPs and PhaFPs were bound to the P(3HB-co-3HA) granules with 3HA (C6–C12) composition of more than 13 mol% in the producing cells, suggesting that localization of these proteins is attributed to the monomer compositions of the copolymers.
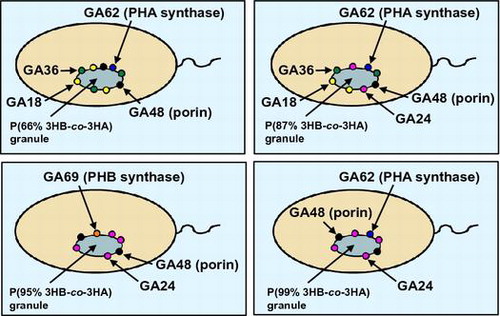
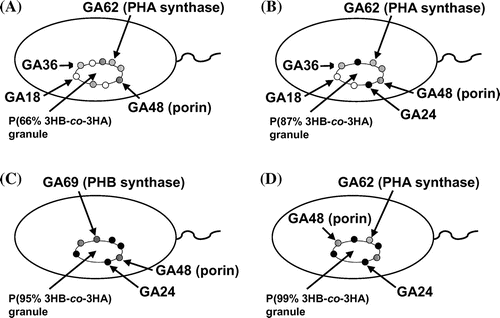
The localization model of the proteins bound to poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) granules with various monomer compositions accumulated in the recombinant strains of Pseudomonas sp. 61-3.
Polyhydroxyalkanoates (PHAs) are accumulated in many bacteria as intracellular carbon and energy storage materials under nutrient-limited conditions with excess carbon.Citation1–3) Within the cells, PHAs are accumulated as granules which contain proteins and lipid involved in their synthesis and regulation.Citation4) There are typically 8–12 granules/cell and the diameter of granules is 0.2 to 0.5 μm in Ralstonia eutropha (formerly Alcaligenes eutrophus).Citation1,5–7) Studies on PHA granules using13C NMR spectroscopy, X-ray diffraction, and electron microscopy have indicated that PHAs are in vivo mobile amorphous and elastomeric state.Citation6–8) To date, some works have revealed the formation mechanism of granules.Citation9–11) Gerngross et al. have reported the localization of the R. eutropha polyhydroxybutyrate (PHB) synthase at the surface of the granules by immunocytochemical methods.Citation12) The granules in the cell are surrounded by a membrane of about 2 nm thickness containing the intracellular enzyme, such as PHA synthase, PHA depolymerase, phasin, and PHA-specific regulator protein.Citation5,13–16) The primary function of phasins, which represent the major components of PHA granule-associated proteins, is to control the surface properties of PHA granules. Phasins strongly bind to the hydrophobic surfaces of growing PHA granules to block the binding of other proteins. The phasins are amphiphilic proteins, which promote PHA biosynthesis and their abundance makes an impact on the PHA granule size.Citation11,17,18) phaP1Re, one of the structural genes of phasin, has been identified in Ralstonia eutropha.Citation17) Overexpression of PhaP1Re results in the formation of many small P(3HB) granules, while the phaP1Re mutant forms only one large P(3HB) granule per cell.Citation17)
Other phaP genes have been identified from Rhodococcus rubber, Acinetobacter sp., Chromatium vinosum, Bacillus megaterium, and Paracoccus denitrificans.Citation19–23) PhaRPd from P. denitrificans is also associated with PHB granules, whose role has been assumed to regulate the expression of phaPPd gene.Citation23) In Pseudomonas oleovorans, two PHA granule-associated proteins, PhaFPo and PhaIPo, have been characterized and the corresponding genes have been cloned and identified.Citation24) Furthermore, PhaFPo negatively regulates the transcription of pha genes, that is, PhaFPo, which is a histone H1-like protein with C-terminal AAKP repeating units, was suggested to repress the expression of phaC1Po and phaIFPo genes.Citation24) These studies have been suggested that the PHA granule-associated proteins not only stabilize PHA granules in the cells, but also regulate the related genes for PHA biosynthesis. On the other hand, in Aeromonas caviae, the phasin (PhaPAc) enhances PHA accumulation and alters P(3HB-co-3-hydroxyhexanoate) copolymer composition,Citation25,26) and it has revealed to function as an activator for A. caviae PHA synthase (PhaCAc).Citation27) Thus, phasins are considered to play a role in PHA biosynthesis, however, the functions have not been fully understood. Therefore, it is important to investigate the roles of phasins and other PHA granule-associated proteins for PHA biosynthesis and the effective production.
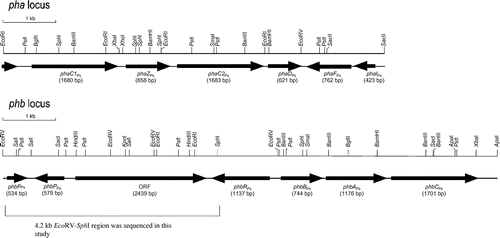
Pseudomonas sp. 61-3 synthesizes two types of PHAs, P(3HB) homopolymer and a random copolymer, P(3HB-co-3HA), consisting of 3-hydroxyalkanoate (3HA) units of 4–12 carbon atoms.Citation28–30) In Pseudomonas sp. 61-3, the study using freeze-fracture electron microscopy revealed that P(3HB) and P(3HB-co-3HA) were accumulated as different granules in the same cell.Citation31) Some proteins associated with the two types of PHA granules in Pseudomonas sp. 61-3 were identified.Citation32) The proteins of 18 kDa (GA18) and 36 kDa (GA36) were specifically bound to P(3HB-co-3HA) granules, whereas the proteins of 24 kDa (GA24) and 48 kDa (GA48, porin) were mainly bound to P(3HB).Citation32) The proteins of 18 and 36 kDa were identified as PhaIPs and PhaFPs, respectively. The amino acid sequences of the proteins showed high homology to those of P. oleovorans, and the two genes were located downstream of the pha locus as described previously (see Fig. ).Citation32) In addition, N-terminal amino acid sequence analyses of the associated proteins with PHA granules and immunoblotting methods revealed that the PHB synthase (PhbCPs) and PHA synthases (PhaC1Ps possibly with PhaC2Ps) from Pseudomonas sp. 61-3 were bound to P(3HB) and P(3HB-co-3HA) granules, respectively.Citation32) However, the reason why their proteins, except PHB and PHA synthases, can bind to each PHA granule has been yet unknown. There are two hypotheses for the reason. One is that the granule-associated proteins (phasins) directly recognize the monomer composition of PHA, and the other is due to the localization caused by the interaction with PHB and/or PHA synthases. In this study, we cloned and identified the gene encoding GA24 (PhbPPs) strongly bound to P(3HB) granule in Pseudomonas sp. 61-3. In addition, we discussed the localization of the proteins associated with P(3HB-co-3HA) granules with various monomer compositions synthesized by the recombinant strains of Pseudomonas sp. 61-3.
Fig. 1. Organization of pha and phb loci in Pseudomonas sp. 61-3.
Material and methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table . Pseudomonas sp. 61-3 and the recombinant strains were grown at 28 °C in NB medium consisting of 1% meat extract (Kyokuto Pharmaceutical Industrial Co., Ltd, Japan), 1% Bactopeptone (Difco, USA) and 0.5% NaCl (pH 7.0). Escherichia coli strains were grown at 37 °C in LB medium.Citation38) When needed, ampicillin (100 mg/L), kanamycin (50 mg/L), tetracycline (12.5 mg/L), and/or gentamicin (10 mg/L) were added to the medium.
Table 1. Bacterial strains and plasmids used in this study.
Production and analysis of PHA
Pseudomonas sp. 61-3 and the recombinant strains were cultivated on a reciprocal shaker (130 strokes/min) at 28 °C for 48 h or 72 h in 500-mL shaking flasks containing 100 mL of a nitrogen-limited MS medium.Citation28,30) Filter-sterilized glucose (2 wt.%) was added to the medium as a sole carbon source. PHA compositions of the isolated granules (see below) were determined by gas chromatography as described previously.Citation28,32)
Isolation of PHA granules and SDS-PAGE analysis
Cells cultivated in MS medium were harvested by centrifugation (7,700 g, 10 min, 4 °C), and then washed twice and resuspended in 2.0 mL of 0.1 M Tris–HCl buffer (pH 7.5). Finally, the cells were disrupted by the treatment of ultrasonication (30 W, 10 s, 20 times). Approximately 1 mL of the broken cell suspension was layered on a discontinuous sucrose gradient from 1 mL each of 2.0, 1.67, 1.33, and 1.0 M sucrose in 0.1 M Tris–HCl buffer (pH 7.5) as described previously.Citation32) After ultracentrifugation (210,000 g, 160 min, 4 °C), the white layers containing PHA granules [P(3HB) and/or P(3HB-co-3HA)] were isolated. The isolated PHA granules were washed twice with 0.1 M Tris-HCl (pH 7.5) by centrifugation (24,000 g, 30 min, 4 °C). Samples of the purified granules were mixed with 2-fold gel buffer (12% of β-mercaptoethanol, 4% of SDS, 20% of glycerol, 0.001% of bromophenol blue, and 0.125 M of Tris-HCl [pH 6.8]), and the proteins were denatured and released from the granules by heating the suspension at 98 °C for 10 min. The proteins were separated by SDS-PAGE with 14% polyacrylamide gels as described by LaemmliCitation39) and stained with a Bio-safe Coomassie (Bio-Rad Laboratories, USA). After SDS-PAGE, the proteins were blotted from the polyacrylamide gels onto PVDF membranes and were identified by N-terminal amino acid sequencing, except PhbCPs was detected by immunoblotting, as described previously.Citation32)
DNA manipulations
Isolation of the total genomic DNA and plasmids, digestion of DNA with restriction endonucleases, agarose gel electrophoresis, and transformation of E. coli were performed by standard procedures.Citation38) The genomic DNA library of Pseudomonas sp. 61-3 was prepared as described previously.Citation30) DNA restriction fragments were extracted from agarose gels using a GENECLEAN Kit (BIO 101, Inc., USA). Conjugation of Pseudomonas sp. 61-3 or the mutant strains with E. coli S17-1 harboring broad-host-range plasmids was performed as described by Friedrich et al.Citation40)
Cloning of the gene encoding the protein associated with P(3HB) granule
For cloning of the gene (phbPPs) encoding the protein (GA24) associated with P(3HB) granule, PCR was performed with primer pairs phbRDS-f1 (5ʹ-TTCCTTGTGAAGGCTCATTGAGGCGTTCAT-3ʹ) and GA24-r1 (5ʹ-TG(T/C)TCIACIG(A/T)IGC(A/G)AA(A/G/T)AT(T/C)TT-3ʹ) using genomic DNA of Pseudomonas sp. 61-3 as a template. The primer phbRDS-f1 was synthesized based on the sequence of the downstream region of phbRPs gene. The degenerate primer GA24-r1 was designed based on the N-terminal amino acid sequence, (M)TFFNLEKLQDAQKANLDLLQ, of GA24 described previously.Citation32) From the information in the nucleotide sequence of the 3-kb amplified fragment, the primers GA24-f2 (5ʹ-AACTTGGAGAAATTGCAAGACGCT-3ʹ) and GA24-f3 (5ʹ-CAACCTAGACCTCCTGCAGCAAAT-3ʹ) were synthesized, and the nested PCR was performed using LA PCRTM in vitro Cloning Kit (TAKARA BIO, Japan) for cloning of the whole phbPPs gene and the downstream region. The nested PCR was firstly performed with primers GA24-f2 and Cassette Primer C1 using Pseudomonas sp. 61–3 genomic DNA digested with SalI as a template. Subsequently, second PCR was performed with primers GA24-f3 and Cassette Primer C2 using the first PCR-amplified product as a template. Finally, the amplified 0.7-kb fragment including phbPPs gene was ligated to pT7Blue T-vector to give pT7-GA24(LA)-F. Furthermore, the 0.5-kb product, including the part of phbPPs gene was amplified by PCR with primer pairs GA24-f3 and GA24-r3 (5’-TTACTTGTTACCGCTTGTTGCCTTGCCAGT-3’) using pT7-GA24(LA)-F as a template and was used for the subsequent hybridization experiments as a probe.
Southern hybridization was performed as described by Southern.Citation41) Preparation of an alkaline phosphatase-labeled probe and detection of hybridization signals on membranes were carried out with Gene Images Alkphos Direct Labelling and Detection System (GE Healthcare, USA). Colony hybridization of genomic DNA libraries of Pseudomonas sp. 61-3 was performed with the probe as described previously.Citation30)
DNA sequencing analysis
DNA fragments to be sequenced were subcloned into pBluescript II KS+. DNA was sequenced by the modified dideoxy-chain termination method basically as described by Sanger et al.Citation42) with a NEN Global Edition IR2 System, LIC4200L (LI-COR, USA). The sequencing reaction was carried out according to the manual supplied with the Thermo Sequenase Cycle Sequencing Kit (USB, USA). The resulting nucleotide sequence was analyzed with SDC-GENETYX information processing software (GENETYX CORPORATION, Japan).
Disruption of phaC1Ps gene
pBSL180 vector was used as an integration to disrupt the chromosomal phaC1Ps gene of Pseudomonas sp. 61-3. pBSEX22 was digested with BglII and EcoRI, and the 1.3-kb BglII-EcoRI fragment (5ʹ- and 3ʹ-truncated phaC1Ps) was then ligated with pBSL180 at the same restriction sites to construct pSLBE13dC1. Conjugation of Pseudomonas sp. 61-3 with E. coli S17-1 (λpir) harboring pSLBE13dC1 was carried out as described previously.Citation33) Southern hybridization analysis was performed using the phaC1Ps gene as a probe to confirm the gene disruption.
Nucleotide sequence accession number
The nucleotide sequence data reported in this study will appear in EMBL, GenBank, and DDBJ database with accession No. LC019127.
Results
Cloning and identification of phbPPs and phbFPs genes
We previously reported that pha and phb loci in Pseudomonas sp. 61-3 involved in the biosyntheses of P(3HB-co-3HA) and P(3HB), respectively.Citation30,32) The genes encoding the P(3HB-co-3HA) granule-associated proteins, GA18 and GA36 were identified as PhaIPs and PhaFPs, and the two genes were located in the downstream region of phaC1ZC2D gene cluster in pha locus.Citation32) Whereas, the gene encoding the P(3HB) granule-associated protein (GA24) remained unidentified although the N-terminal amino acid sequence was determined.
It was reported that Azotobacter vinelandii UW136 had a P(3HB) biosynthetic gene cluster (phbRBAC) as same as phb locus in Pseudomonas sp. 61-3,Citation43) and phbPAv gene encoding a putative P(3HB) granule-associated protein phasin was present downstream of phbRAv in the same orientation. Similarly, the region downstream of phbRPs gene of Pseudomonas sp. 61-3 was explored to find GA24 gene (phbPPs). Firstly, we attempted PCR with primer pairs phbRDS-f1, which corresponded to the downstream sequence of phbRPs gene, and GA24-r1, which was a degenerate primer based on the N-terminal amino acid sequence of GA24, using genomic DNA of Pseudomonas sp. 61-3 as a template. As a result, approximately 3-kb DNA fragment was amplified by PCR. From the information in the nucleotide sequence of the PCR product, furthermore, nested PCR and colony hybridization with genomic DNA library of Pseudomonas sp. 61-3 were performed with the probe, including the gene encoding GA24 (PhbPPs) for cloning of the downstream region of phbPPs gene as described in materials and methods section. A positive clone isolated by colony hybridization was used for southern hybridization analysis. The positive 7.6-kb HindIII and 6.9-kb SacI fragments were cloned into pBluescript II KS+ and partially sequenced. From the information in the nucleotide sequences obtained from the positive clones and the nucleotide sequence of the 3-kb PCR product amplified with primers phbRDS-f1 and GA24-r1 as described above, the 4.2 kb EcoRV-SphI region downstream of phbRPs gene was completely sequenced. The restriction maps of PHA biosynthesis genes (pha and phb loci), which have been so far elucidated, are shown in Fig. . In the downstream region of phbRPs gene, three potential ORFs were identified by computer analysis. The nucleotide sequence revealed homologies to genes encoding phasin GA24 (PhbPPs) and the transcriptional negative regulator (PhbFPs) in A. vinelandii UW136 and Azotobacter sp. FA8.Citation43,44) phbPPs and phbFPs encoded putative proteins composed of 192 amino acids with a calculated molecular mass of 20.4 kDa and 177 amino acids with a calculated molecular mass of 19.6 kDa, respectively. The deduced amino acid sequence of PhbPPs (GA24) showed high homologies to PhaP of Azotobacter sp. FA8 (57% identity)Citation44) and PhbP of A. vinelandii AvOP (54% identity).Citation43) Furthermore, the deduced amino acid sequence of PhbFPs revealed high homologies to PhaF of Azotobacter sp. FA8 (69% identity)Citation44) and PhbF of A. vinelandii AvOP (68% identity).Citation43) PhbPPs (GA24) of Pseudomonas sp. 61-3 was expected to have a function of phasin protein, such as reported elsewhere.Citation11,17,18) PhbFPs also showed 37.5% identity of amino acid homology to PhaR of P. denitrificans.Citation23) The role of PhaR protein in P. denitrificans is assumed to negatively regulate the transcriptional expression of phaP gene. In addition, Pfam program showed that PhbFPs comprised three domains, PHB/PHA accumulation regulator DNA-binding domain (amino acid positions 10 to 73), PHB accumulation regulatory domain (amino acid positions 75 to 114), and PHB accumulation regulatory domain (amino acid positions 116 to 154). Therefore, PhbFPs of Pseudomonas sp. 61-3 is probably a negative transcriptional regulator to repress the expression of phbPPs gene.
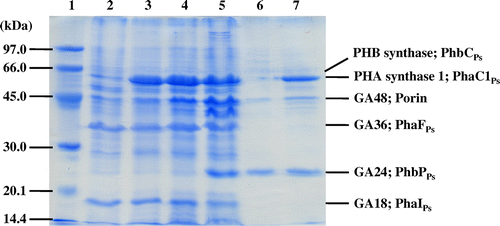
Fig. 2. SDS-PAGE analysis of native PHA granules isolated from the recombinant strains of Pseudomonas sp. 61–3. Lane 1, molecular weight markers; lane 2, Pseudomonas sp. 61–3 (phbC::tet); lane 3, Pseudomonas sp. 61–3 (phbC::tet)/pJASc22; lane 4, Pseudomonas sp. 61–3 (phbC::tet)/pJKSc46-pha; lane 5, Pseudomonas sp. 61–3 (phbC::tet)/pJKSc54-phab; lane 6, Pseudomonas sp. AC1-TnK; and lane 7, Pseudomonas sp. BCG-TcGm/pJKSc54-phab.
Interestingly, another ORF was found between phbPPs and phbRPs genes in phb locus of Pseudomonas sp. 61-3. Such ORF has not been found in the P(3HB) biosynthesis gene clusters of A. vinerandii UW136 and Azotobacter sp. FA8 (Fig. ).Citation43,44) The ORF encoded a putative protein composed of 812 amino acids with a calculated molecular mass of 90.2 kDa. The deduced amino acid sequence of the ORF showed high homologies to putative poly(3-hydroxyalkanoate) synthetases of Pseudomonas sp. GM48 (92% identity, accession No. WP_007988013) and P. putida (87% identity, accession No. WP_033040191), and putative poly(3-hydroxybutyrate) depolymerase of A. vinelandii DJ (59% identity) according to the genomic informations. Pfam program showed that the function (DUF3141) of these putative proteins having α/β hydrolase domain was unknown. Function of the ORF found in this study remains unknown although the possibility of PHA synthase or PHA depolymerase have been investigated in vivo (data not shown).
Analysis of PHA granules and localization of PHA granule-associated proteins
phbCPs- or phaC1Ps-disrupted strains of Pseudomonas sp. 61-3 was used as a host in order to synthesize the only one type of PHA, since the wild-type strain accumulated two types of PHAs, P(3HB) homopolymer and P(3HB-co-3HA) copolymer in the same cells (Fig. ).Citation30,31,37) The P(3HB-co-3HA) granules with various monomer compositions were synthesized by the recombinant strains of Pseudomonas sp. 61-3, and PHA granules accumulated in the cells were isolated by sonication and a subsequent sucrose density gradient method as described in materials and methods section. Only one white band was observed at the interfaces of 0–1.0 M or 1.3–1.67 sucrose from each of the cell extracts of all recombinant strains of Pseudomonas sp. 61-3, indicating that only one type of PHA is synthesized in the cells. P(3HB-co-3HA) granules with relatively low 3HB compositions (less than 66 mol%) were collected at the interface of 0–1.0 M sucrose, and the copolymer granules with high 3HB compositions (more than 87 mol%) were collected at the interface of 1.33–1.67 M sucrose. The monomer compositions of the copolymer granules accumulated by the recombinant strains of Pseudomonas sp. 61-3 were determined by gas chromatography (Table ) and the proteins associated with the granules were separated by SDS-PAGE (Fig. ). The relationship between the monomer compositions of the isolated PHA granules and the proteins bound to the respective PHA granules is shown in Table .
Table 2. Relationship of the monomer composition of PHA accumulated by recombinant strains of Pseudomonas sp. 61-3 and the granule-associated proteins.
In Fig. , the 60–70 kDa proteins were identified as PHB synthase (PhbCPs) or PHA synthase 1 (PhaC1Ps) from Pseudomonas sp. 61-3 by analyses of the N-terminal amino acid sequences and immunoblotting as described previously.Citation32) Whereas, PHA synthase 2 (PhaC2Ps) of Pseudomonas sp. 61-3 was not able to be detected from P(3HB-co-3HA) granules, suggesting that PhaC1Ps was the major PHA providing enzyme in Pseudomonas sp. 61-3 as described previously.Citation32) GA18, GA36, and GA48 proteins were also identified as PhaIPs, PhaFPs, and porin D, respectively.Citation32) As shown in Fig. and Table , PhaC1Ps was detected with the PHA granules isolated from five strains except Pseudomonas sp. AC1-TnK, and PhbCPs was weakly detected with the PHA granule in Pseudomonas sp. AC1-TnK. PhaIPs and PhaFPs were detected with the PHA granules isolated from Pseudomonas sp. 61-3 (phbC::tet) and the recombinant strains harboring pJASc22, pJKSc46-pha, and pJKSc54-phab. GA24 identified as PhbPPs in this study was confirmed with the PHA granules isolated from Pseudomonas sp. 61-3 (phbC::tet)/pJKSc54-phab, Pseudomonas sp. AC1-TnK, and Pseudomonas sp. BCG-TcGm/pJKSc54-phab. However, PhaIPs and PhaFPs were not able to be associated with the granules in Pseudomonas sp. AC1-TnK and Pseudomonas sp. BCG-TcGm/pJKSc54-phab. In other words, PhbPPs (GA24) could bind to P(3HB-co-3HA) granules with 3HB composition of more than 87 mol%, but could not be almost bind to the copolymer granule with 3HB composition of less than at least 66 mol%. Whereas, PhaIPs (GA18) and PhaFPs (GA36) could bind to P(3HB-co-3HA) granules with 3HA (C6-C12) composition of more than 13 mol%. GA48 (porin) was considered to be nonspecifically bound to all PHAs regardless of the monomer compositions. Interestingly, the all phasin or phasin-like proteins (PhbPPs, PhaIPs, and PhaFPs) were associated with P(87% 3HB-co-13% 3HA) granule.
PhbPPs (GA24) was firstly found as the protein associated with P(3HB) granule, which was one of the two types of PHAs, P(3HB), and P(3HB-co-3HA), accumulated in Pseudomonas sp. 61-3. In addition, PhbPPs could bind to P(3HB-co-3HA) granules obtained from the cells of Pseudomonas sp. 61-3 (phbC::tet)/pJKSc54-phab and Pseudomonas sp. BCG-TcGm/pJKSc54-phab, although the phbCPs gene of these strains was disrupted. Therefore, PhbPPs is likely to recognize the monomer units, probably 3HB unit, of copolymers without interaction of PHB synthase.
Discussion
Pseudomonas sp. 61-3 produces two types of PHAs, P(3HB) homopolymer and P(3HB-co-3HA) random copolymer and accumulates them as different granules in the cell.Citation28,29,31) The genes involved in P(3HB) and P(3HB-co-3HA) biosyntheses from Pseudomonas sp. 61-3 were cloned and identified previously.Citation30,32,33) In the previous report, two PHA granules, P(3HB) and P(3HB-co-3HA), were isolated from Pseudomonas sp. 61-3, and polyester synthases (PhaC1Ps and PhbCPs) and the proteins (PhaIPs and PhaFPs) associated with PHA granules were identified.Citation32) In this report, another protein (GA24) associated with P(3HB) granule was identified. The deduced amino acid sequence of GA24 gene revealed high homologies to those of PhaPAs of Azotobacter sp. FA8Citation44) and PhbPAv of A. vinelandii AvOP.Citation43) Therefore, GA24 was referred to as PhbPPs, and it was probably anticipated to stabilize the granules of P(3HB) and P(3HB-co-3HA) with high 3HB fraction (more than 87 mol%) in the producing cells as a phasin. In this experiment, we also found a potential ORF downstream of phbPPs gene in the opposite direction (Fig. ). The putative translational product of the ORF revealed high homologies to PhaFAs of Azotobacter sp. FA8Citation44) and to PhbFAv of A. vinelandii AvOP.Citation43) Therefore, the ORF was referred to as phbFPs, and PhbFPs was assumed to repress the transcriptional expression of phbPPs gene in Pseudomonas sp. 61-3, since it also showed 37.5 and 56% identities to PhaRPd of P. denitrificans and PhaRRe of R. eutropha, respectively, which were known as transcriptional repressors to regulate the expression of phasins.Citation23,45,46) Moreover, another large ORF was found between phbPPs and phbRPs genes in phb locus of Pseudomonas sp. 61-3 (Fig. ). The deduced amino acid sequence of the ORF showed high homologies to putative poly(3-hydroxyalkanoate) synthetases of Pseudomonas sp. GM48 (92% identity) and P. putida (87% identity), and a putative poly(3-hydroxybutyrate) depolymerase of A. vinelandii DJ (59% identity) according to the genomic informations. Additionally, the transcription of the ORF was confirmed by semi-quantitative RT-PCR (data not shown). Therefore, it was expected that the ORF might encode PHA synthase or PHA depolymerase, especially for biosynthesis or degradation of P(3HB), since the ORF was also found in phb locus of Pseudomonas sp. 61-3. However, the function of the ORF remains unknown, and we will investigate and report it in the next research.
In this study, localization of the proteins (PhbPPs, PhaIPs, and PhaFPs) associated with the granules of P(3HB) and P(3HB-co-3HA) in the cells of Pseudomonas sp. 61-3 was supposed to be attributed to the monomer compositions of polymers. Our findings are summarized and depicted in Fig. . PhbPPs (GA24) was detected with the granules of P(3HB-co-3HA) copolymers with 3HB composition of more than 87 mol%, and both PhaIPs (GA18) and PhaFPs (GA36) were detected with the granules of the copolymers with 3HA (C6-C12) composition of more than 13 mol% (Table and Fig. ). PhbPPs was detected from the polyester granules in the cells of Pseudomonas sp. 61-3 (phbC::tet)/pJKSc54-phab and Pseudomonas sp. BCG-TcGm/pJKSc54-phab whose strains were phbCPs-disruptants. PhaIPs and PhaFPs were detected from the polyester granules in the cells of Pseudomonas sp. 61-3 (phbC::tet), Pseudomonas sp. 61-3 (phbC::tet)/pJASc22, Pseudomonas sp. 61-3 (phbC::tet)/pJKSc46-pha, and Pseudomonas sp. 61-3 (phbC::tet)/pJKSc54-phab, whereas these proteins could not be confirmed on the surface of the polyester granules in the cells of Pseudomonas sp. BCG-TcGm/pJKSc54-phab where phaC1Ps gene was introduced. Thus, the three granule-associated proteins, PhbPPs (GA24), PhaIPs (GA18), and PhaFPs (GA36), appear to recognize the polyester chain directly without interaction and support of polyester synthases. Additional copies of phbPPs, phaIPs, and/or phaFPs genes did not affect the production and the monomer compositions of P(3HB-co-3HA) copolymers synthesized by the recombinant strains of Pseudomonas sp. 61-3 (phbC::tet) as a host (data not shown), unlike phaPAc of A. caviae.Citation25–27) It has been reported that PhaPAc activate A. caviae PHA synthase (PhaCAc), but not R. eutropha PHA synthase (PhaCRe).Citation27) This may be due to the low amino acid sequence identity (13%) between PhaPAc and PhaP1Re of R. eutropha. Similarly, the PhbPPs shows a low identity (23%) to PhaPAc. Also, the PHA granule-associated proteins of Pseudomonas sp. 61-3 may form multimer. For example, PhaPs of Aeromonas hydrophila and R. eutropha have been reported to form trimers and tetramers by X-ray analysis, respectively.Citation11,47) However, the relationship between the multimeric form of phasins and the binding to PHA granule has not been elucidated yet.
Fig. 3. The localization model of the proteins associated with polyester granules accumulated in (A) Pseudomonas sp. 61–3 (phbC::tet)/pJKSc46-pha, (B) Pseudomonas sp. 61–3 (phbC::tet)/pJKSc54-phab, (C) Pseudomonas sp. AC1-TnK, and (D) Pseudomonas sp. BCG-TcGm/pJKSc54-phab.
In vivo, PHAs are mobile amorphous and elastomeric state, and PHA granules are surrounded by a membrane. Several works have been carried out to reveal the forming mechanism of membrane, and some models have been proposed.Citation4,6–8,15,20) According to the first model, PHA granules are surrounded by a phospholipid membrane with embedded proteins consisting of PHA synthase, intracellular PHA depolymerase, phasin protein, and other proteins.Citation4,20) The second model has been proposed that PHA granule-associated proteins present on the phospholipid monolayer.Citation15) The third model has been proposed that PHA granule-associated proteins present on a much more membrane structure with phospholipid bilayer.Citation15,48) In Pseudomonas sp. 61-3, the membrane structure surrounded PHA granules has not been elucidated, however, it was found that PhbPPs (GA24), PhaIPs (GA18), and PhaFPs (GA36) specifically bound to P(3HB) and P(3HB-co-3HA) granules, respectively, in the previous study.Citation32) In addition, our data suggest that binding of their proteins to PHA granules would be due to recognizing the monomer units of polymers by the proteins. While, Mayer et al. have reported that the boundary layer structure of PHA granules in bacteria might vary by PHA monomer composition.Citation49) Therefore, the specific binding of PhbPPs, PhaIPs, and PhaFPs to PHA granules might be due to the difference of boundary layer structure of surrounded PHAs. Possibly, the layer structure might be attributed to the monomer compositions of P(3HB-co-3HA) copolymers. PHB/PHA synthases, PHB/PHA depolymerases, phasins, and regulatory proteins are known as major PHA granule-associated proteins. These proteins are definitely important for biosynthesis and/or degradation of PHAs. The findings and the observations obtained here will lead to the effective production and the biosynthesis of PHAs, P(3HB-co-3HA) copolymers, with favorable monomer compositions. For example, the P(94% 3HB-co-6% 3HA) copolymer synthesized by the recombinant strain of Pseudomonas sp. 61-3 is known to have properties similar to low-density polyethylene (LDPE), and the copolymer is a practical material for application of biodegradable plastics.Citation37) Thus, to produce practical P(3HB-co-3HA) copolymer with high 3HB fraction effectively, sufficient amounts of amphiphilic PhbPPs might have to be provided in the producing cells for stabilization of hydrophobic PHA granules in the cells. Therefore, further studies on PHA biosynthesis and PHA granule formation in Pseudomonas sp. 61-3 is in progress.
Acknowledgment
We are grateful to Dr Ken’ichiro Matsumoto for the technical assistance and to NBRP (National BioResource Project, Japan) for the plasmid pBSL180 and E. coli S17-1 (λpir).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990;54:450–472.
- Müller HM, Seebach D. Poly(hydroxyalkanoates): a fifth class of physiologically important organic biopolymers? Angew. Chem. Int. Ed. Engl. 1993;32:477–502.10.1002/(ISSN)1521-3773
- Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 1999;63:21–53.
- Griebel R, Smith Z, Merrick JM. Metabolism of poly-β-hydroxybutyrate. I. Purification, composition, and properties of native poly-β-hydroxybutyrate granules from Bacillus megaterium. Biochemistry. 1968;7:3676–3681.10.1021/bi00850a047
- Lundgren DG, Pfister RM, Merrick JM. Structure of poly-β-hydroxybutyric acid granules. J. Gen. Microbiol. 1964;34:441–446.10.1099/00221287-34-3-441
- Barnard GN, Sanders JK. The poly-β-hydroxybutyrate granule in vivo. A new insight based on NMR spectroscopy of whole cells. J. Biol. Chem. 1989;264:3286–3291.
- Kawaguchi Y, Doi Y. Structure of native poly(3-hydroxybutyrate) granules characterized by X-ray diffraction. FEMS Microbiol. Lett. 1990;70:151–155.10.1111/fml.1990.70.issue-2
- Horowitz DM, Sanders JKM. Biomimetic, amorphous granules of polyhydroxyalkanoates: composition, mobility, and stabilization in vitro by proteins. Can. J. Microbiol. 1995;41:115–123.10.1139/m95-177
- Fuller RC. Microbial inclusions with special reference to PHA inclusions and intracellular boundary envelopes. Int. J. Biol. Macromol. 1999;25:21–29.10.1016/S0141-8130(99)00011-2
- Sudesh K, Abe H, Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog. Polym. Sci. 2000;25:1503–1555.
- Neumann L, Spinozzi F, Sinibaldi R, Rustichelli F, Pötter M, Steinbüchel A. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 2008;190:2911–2919.10.1128/JB.01486-07
- Gerngross TU, Reilly P, Stubbe J, Sinskey AJ, Peoples OP. Immunocytochemical analysis of poly-β-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: localization of the synthase enzyme at the surface of PHB granules. J. Bacteriol. 1993;175:5289–5293.
- Merrick JM, Lundgren DG, Pfister RM. Morphological changes in poly-β-hydroxybutyrate granules associated with decreased susceptibility to enzymatic hydrolysis. J. Bacteriol. 1965;89:234–239.
- Merrick JM. Effect of polymyxin B, tyrocidine, gramicidin D, and other antibiotics on the enzymatic hydrolysis of poly-β-hydroxybutyrate. J. Bacteriol. 1965;90:965–969.
- Mayer F, Hoppert M. Determination of the thickness of the boundary layer surrounding bacterial PHA inclusion bodies, and implications for models describing the molecular architecture of this layer. J. Basic Microbiol. 1997;37:45–52.
- Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem. J. 2003;376:15–33.10.1042/BJ20031254
- Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 1995;177:2425–2435.
- York GM, Junker BH, Stubbe JA, Sinskey AJ. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 2001;183:4217–4226.10.1128/JB.183.14.4217-4226.2001
- Liebergesell M, Steinbüchel A. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur. J. Biochem. 1992;209:135–150.10.1111/ejb.1992.209.issue-1
- Pieoer-Fürst U, Madkour MH, Mayer F, Steinbüchel A. Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J. Bacteriol. 1994;176:4328–4337.
- Schembri MA, Woods AA, Bayly RC, Davies JK. Identification of a 13-kDa protein associated with the polyhydroxyalkanoic acid granules from Acinetobacter spp. FEMS. Microbiol. Lett. 1995;133:277–283.10.1111/fml.1995.133.issue-3
- McCool GJ, Cannon MC. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 1999;181:585–592.
- Maehara A, Ueda S, Nakano H, Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 1999;181:2914–2921.
- Prieto MA, Bühler B, Jung K, Witholt B, Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 1999;181:858–868.
- Kichise T, Fukui T, Yoshida Y, Doi Y. Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int. J. Biol. Macromol. 1999;25:69–77.
- Fukui T, Kichise T, Iwata T, Doi Y. Characterization of 13 kDa granule-associated protein in Aeromonas caviae and biosynthesis of polyhydroxyalkanoates with altered molar composition by recombinant bacteria. Biomacromolecules. 2001;2:148–153.
- Ushimaru K, Motoda Y, Numata K, Tsuge T. Phasin proteins activate Aeromonas caviae polyhydroxyalkanoate (PHA) synthase but not Ralstonia eutropha PHA synthase. Appl. Environ. Microbiol. 2014;80:2867–2873.10.1128/AEM.04179-13
- Kato M, Bao HJ, Kang CK, Fukui T, Doi Y. Production of a novel copolyester of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl. Microbiol. Biotechnol. 1996;45:363–370.
- Kato M, Fukui T, Doi Y. Biosynthesis of polyester blends by Pseudomonas sp. 61-3 from alkanoic acids. Bull. Chem. Soc. Jpn. 1996;69:515–520.
- Matsusaki H, Manji S, Taguchi K, Kato M, Fukui T, Doi Y. Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. strain 61-3. J. Bacteriol. 1998;180:6459–6467.
- Fukui T, Kato M, Matsusaki H, Iwata T, Doi Y. Morphological and 13C-nuclear magnetic resonance studies for polyhydroxyalkanoate biosynthesis in Pseudomonas sp. 61-3. FEMS Microbiol. Lett. 1998;164:219–225.
- Matsumoto K, Matsusaki H, Taguchi K, Seki M, Doi Y. Isolation and characterization of polyhydroxyalkanoates inclusions and their associated proteins in Pseudomonas sp. strain 61-3. Biomacromolecules. 2002;3:787–792.
- Matsumoto K, Matsusaki H, Taguchi S, Seki M, Doi Y. Cloning and characterization of the Pseudomonas sp. 61-3 phaG gene involved in polyhydroxyalkanoate biosynthesis. Biomacromolecules. 2001;2:142–147.
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology. 1983;1:784–791.10.1038/nbt1183-784
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene. 1995;160:59–62.10.1016/0378-1119(95)00141-R
- Matsusaki H, Abe H, Taguchi K, Fukui T, Doi Y. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant bacteria expressing the PHA synthase gene phaC1 from Pseudomonas sp. 61-3. Appl. Microbiol. Biotechnol. 2000;53:401–409.
- Matsusaki H, Abe H, Doi Y. Biosynthesis and properties of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant strains of Pseudomonas sp. 61-3. Biomacromolecules. 2000;1:17–22.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, (NY): Cold Spring Harbor Laboratory Press; 1989.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685.10.1038/227680a0
- Friedrich B, Hogrefe C, Schlegel HG. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 1981;147:198–205.
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517.10.1016/S0022-2836(75)80083-0
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Nat. Acad. Sci. USA. 1977;74:5463–5467.10.1073/pnas.74.12.5463
- Peralta-Gil M, Segura D, Guzman J, Servin-Gonzalez L, Espin G. Expression of the Azotobacter vinelandii poly-β-hydroxybutyrate biosynthetic phbBAC operon is driven by two overlapping promoters and is dependent on the transcriptional activator PhbR. J. Bacteriol. 2002;184:5672–5677.10.1128/JB.184.20.5672-5677.2002
- Pettinari MJ, Chaneton L, Vazquez G, Steinbüchel A, Méndez BS. Insertion sequence-like elements associated with putative polyhydroxybutyrate regulatory genes in Azotobacter sp. FA8. Plasmid. 2003;50:36–44.10.1016/S0147-619X(03)00009-X
- Pötter M, Madkour MH, Mayer F, Steinbüchel A. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology. 2002;148:2413–2426.
- Pötter M, Müller H, Steinbüchel A. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology. 2005;151:825–833.10.1099/mic.0.27613-0
- Zhao M, Li Z, Zheng W, Lou Z, Chen GQ. Crystallization and initial X-ray analysis of polyhydroxyalkanoate granule-associated protein from Aeromonas hydrophila. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2006;62:814–819.10.1107/S1744309106025000
- Stuart ES, Fuller RC, Lenz RW. The ordered macromolecular surface of polyester inclusion bodies in Pseudomonas oleovotans. Can. J. Microbiol. 1995;41:84–93.10.1139/m95-174
- Mayer F, Madkour MH, Pieper-Fürst U, Wieczorek R, Liebergesell M, Steinbüchel A. Electron microscopic observations on the macromolecular organization of the boundary layer of bacterial PHA inclusion bodies. J. Gen. Appl. Microbiol. 1996;42:445–455.10.2323/jgam.42.445
