Abstract
This study presents the first analysis of expressed transcripts in the spermary and ovary of Hyriopsis schlegelii (H. schlegelii). A total of 132,055 unigenes were obtained and 31,781 of these genes were annotated. In addition, 19,511 upregulated and 25,911 downregulated unigenes were identified in the spermary. Ten sex-determination genes were selected and further analyzed by real-time PCR. In addition, mammalian genes reported to govern sex-determination pathways, including Sry, Dmrt1, Dmrt2, Sox9, GATA4, and WT1 in males and Wnt4, Rspo1, Foxl2, and β-catenin in females, were also identified in H. schlegelii. These results suggest that H. schlegelii and mammals use similar gene regulatory mechanisms to control sex determination. Moreover, genes associated with dosage compensation mechanisms, such as Msl1, Msl2, and Msl3, and hermaphrodite phenotypes, such as Tra-1, Tra-2α, Tra-2β, Fem1A, Fem1B, and Fem1C, were also identified in H. schlegelii. The identification of these genes indicates that diverse regulatory mechanisms regulate sexual polymorphism in H. schlegelii.
Graphical abstract
19,511 upregulated and 25,911 downregulated genes were detected in the spermary compared to the ovary, including many sex determination and differentiation genes.
The variety of sex-determination pathways among phyla may reflect the rapid evolution of sex-related genes. Many sex-determination genes, including Sry (sex-determination region of chromosome Y),Citation1) Sox9 (SRY box-containing gene 9),Citation2) Dmrt1 (Doublesex and mab3-related transcription factor 1) and Dmrt2 (Doublesex and mab3-related transcription factor 2),Citation3) Foxl2 (forkhead box L2),4) Rspo1 (R-spondin1), and β-catenin,Citation5) have been implicated in sexual determination. Dmrt genes have been identified in species such as mammals,Citation6) birds,Citation7) amphibians,Citation8) and teleosts.Citation9) However, Sry genes have not been identified in non-mammalian vertebrates. In male sex-determination pathways, sex-determination genes, including Sry, Dmrt1, and Sox9, induce testis differentiation.Citation10,Citation11) In bipotential, undifferentiated gonads, Sox9 induces testis differentiation gene expression and promotes Sertoli cell differentiation.Citation12) There are two independent sex-determination pathways in female development: the Foxl2 transcription factor pathway and the Rspo1/β-catenin pathway.4,Citation5) In mature female Foxl2 gene knockout mice, ovaries differentiate into testes.4) In somatic cells, Rspo1 secretion is required for ovarian differentiation because this protein activates the WNT/β-catenin signaling pathway in female sex development.Citation5)
Studies of sex-determination mechanisms in invertebrates have primarily focused on insects and nematodes. The Doublesex gene was identified in the fruit fly Drosophila melanogaster (D. melanogaster) as a major sex regulatory gene,Citation13) and the Mab-3 gene was identified in the nematode Caenorhabditis elegans (C. elegans).Citation14) The Doublesex and Mab-3 genes are required for male sexual development in D. melanogaster and C. elegans, respectively. Two gene families, Fem (feminization) and Tra, have been characterized in nematodes.Citation15,Citation16) The activation of Tra-1 (transformer 1) determines female fate via the suppression of male development genes, while Tra-2 (transformer 2) controls the activity of Tra-1 via the inhibition of Fem genes.Citation15) However, the sex-determination regulatory factors and signal pathways in freshwater bivalves have not been characterized.
The transcriptome represents the total set of RNA transcripts, primarily mRNA and non-coding RNA, in the cell in specific functional states. Transcriptome sequencing permits the rapid and complete elucidation of transcript sequence information in certain tissues or organs. During the 26- to 32-month sexual maturity process, a unique hermaphroditic phenomenon occurs in the spermary of the freshwater pearl mussel Hyriopsis schlegelii (H. schlegelii).Citation17) To obtain more information about sex-determination genes and regulation mechanisms, a high-throughput sequencing technology was used to characterize the spermary and ovary transcriptomes in H. schlegelii. A number of important candidate sex-determination genes were screened. Ten genes related to sex determination were then selected for differential expression analysis in the spermary and ovary using real-time PCR. These results may provide valuable information about the genes and proteins associated with sex determination and the hermaphroditic phenomenon in freshwater bivalves.
Materials and methods
Animal’s, RNA isolation and sequencing
Healthy adult H. schlegelii approximately four years of age were obtained from the Fuzhou Hongmen Reservoir Exploitation Company in Fuzhou City, Jiangxi Province, China. Permission to use H. schlegelii for these experiments was obtained from the Wildlife Protection Center of Nanchang University, Jiangxi, China. Total RNA was extracted using 1 mL of Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After treatment with RNase-free DNase I (Promega), two RNA pools, one from four-year-old spermary and one from four-year-old ovary tissues, were submitted to BGI (Shenzhen, Guangdong, China), where two cDNA libraries were prepared and Solexa sequencing was performed.
Output statistics and assembly
Raw reads were obtained from a sequencing machine. Sequence reads containing sequencing adapters and low-quality sequence reads were removed, and clean reads were generated after filtering the raw reads. Transcriptome de novo assembly was performed using the short reads assembly program Trinity.18) Trinity combines reads with certain lengths of overlap to form longer fragments called contigs. The contigs are subsequently connected to obtain sequences that cannot be extended on either end, defined as unigenes.
Unigene function annotation and classification.
Using Blastx, unigene sequences were aligned to protein databases, such as nr, Swiss-Prot, KEGG, and COG (E-value < 0.00001), and Blastn was used to align the unigene sequences to the nucleotide database nt (E-value < 0.00001). The Blast2GO programCitation19) was used to obtain GO annotations for the unigenes. After obtaining GO annotations for every unigene, we used WEGO software20) to conduct a GO functional classification for all unigenes and determine the distribution of gene functions within species.
Unigene expression difference analysis
Fragments per kb per million fragments was used to calculate unigene expression.Citation21) The differentially expressed genes (DEGs) between the spermary and ovary were identified using a previously described protocol.Citation22) The FDR (false discovery rate) control is a statistical method used in multiple hypothesis testing to correct for p-value. In the analyses conducted herein, we applied a threshold of FDR ≤ 0.001 and an absolute value of log2Ratio ≥ 1 to determine the significance of gene expression differences.
RT-qPCR
The RNA samples used for real-time quantitative PCR (RT-qPCR) were the same as those used for the spermary and ovary DGE experiments. Total RNA was treated with RNase-free DNase I (Promega, USA) to eliminate genomic DNA contamination.Citation23,Citation24) cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Promega, USA). RT-qPCR was performed using a real-time PCR SYBR Green detection system (Eppendorf). Each cDNA was analyzed in quadruplicate, and the average threshold cycle (Ct) was calculated for each sample. The relative expression levels were calculated using the 2-△△CT method,Citation25) and the average Ct was analyzed for all genes to correct for differences in the cDNA input. All reactions were performed in duplicate. The β-actin gene was used as a reference for normalization. The forward and reverse primer sequences are provided in Table .
Table 1. Sequences of the primers used in this study.
Results
Output statistics and assembly results
To obtain an overview of the gene expression profile in H. schlegelii sex determination, normalized spermary and ovary cDNA libraries were sequenced. A total of 51.7 million and 51.4 million clean reads with 97.36 and 97.67% Q20 bases (base quality greater than 20) were generated from the spermary and ovary cDNA libraries, respectively, and a total of 279,227 and 228,674 contigs with average lengths of 270 and 281 bp, respectively, were assembled based on high-quality reads (Table ). After further data processing, 142,717 and 117,293 unigenes with average lengths of 472 and 484 bp, respectively, were obtained. In total, 132,055 non-redundant unigenes with an average length of 603 bp were obtained.
Table 2. Overview of sequencing and assembly.
Non-redundant and COG classification
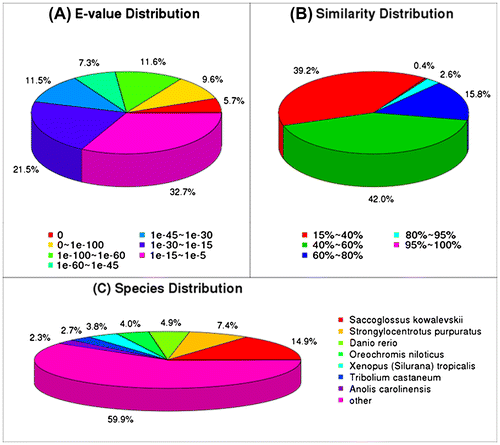
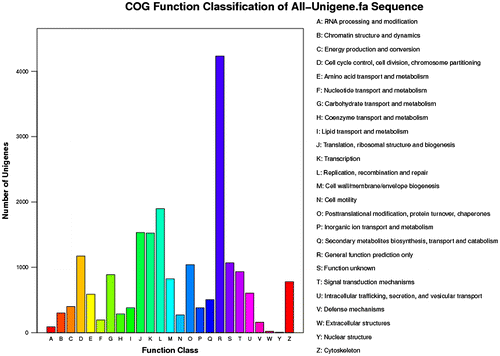
In the present study, 132,055 unigenes were searched using BlastX alignment against the non-redundant database, and 28,139 unigenes were annotated. The E-value distribution of the top hits revealed that 26.9% of the annotated sequences had high homology (E-value less than 1.0E-60), while 73.1% of the sequences had E-values ranging from 1.0E-5 to 1.0E-60 (Fig. (A)). The similarity distribution revealed that 18.8% of the sequences had similarities of greater than 60%, whereas 81.2% of the sequences exhibited similarity ranging from 15 to 60% (Fig. (B)). Among the sequences, 14.9% yielded top matches with sequences from Saccoglossus kowalevskii, followed by Strongylocentrotus purpuratus (7.4%), Danio rerio (4.9%) Oreochromis niloticus (4.0%), Xenopus (silurana) tropicalis (3.8%), Tribolium castaneum (2.7%), and Anolis carolinensis (2.3%) (Fig. (C)). Among the 25 COG categories, the “General function prediction” (4231), “Replication, recombination and repair” (1898), “Translation, ribosomal structure and biogenesis” (1531), and “Transcription” (1524) groups were dominant (Fig. ).
Gene ontology classification
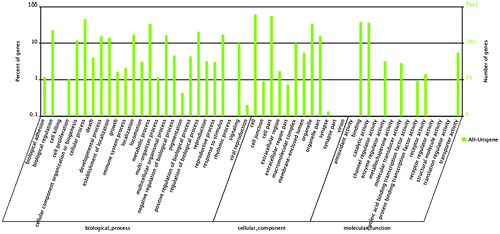
Gene ontology (GO) assignments were used to classify the functions of the predicted H. schlegelii genes. Among the 41 GO classification functional groups, “cell” represented the largest term (4542), followed by “cellular process” (3337) and “binding” (2819). We identified categories involving reproduction, such as “reproduction” (239), “reproductive process” (227), and “viral reproduction” (15) (Fig. ).
Fig. 3. GO classification of unigenes.
Transcript differences between the ovary and spermary
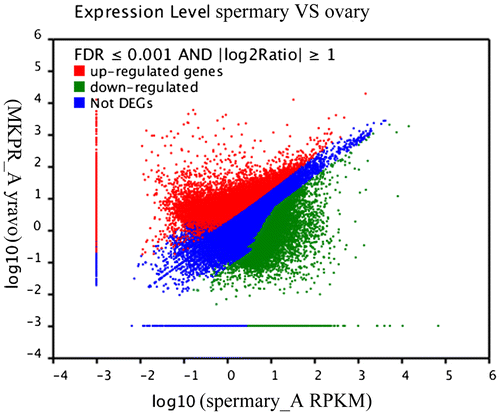
Differential expression between the ovary and spermary libraries was analyzed to identify the genes involved in sex determination and differentiation. Based on the applied criteria (twofold or greater change and p < 0.001), 45,422 unigenes were identified as differentially expressed between the ovary and spermary, comprising 19,511 upregulated unigenes and 25,911 downregulated unigenes in the spermary (Fig. ).
Expression patterns of 10 important transcripts related to sex determination
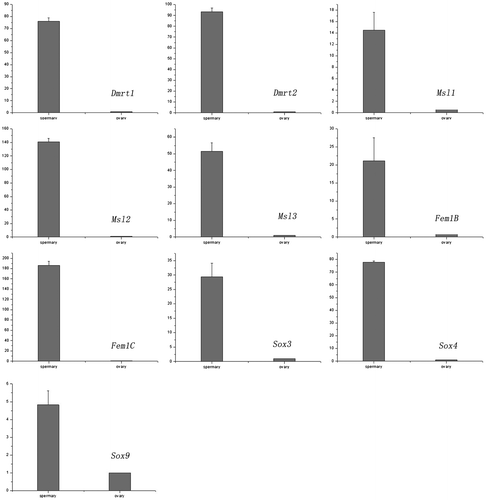
We analyzed the expression patterns of 10 transcripts between the spermary and ovary to investigate the potential roles of these genes in sexual differentiation and determination. All of the selected genes associated with sex determination and differentiation were highly expressed in the spermary compared with the ovary (Fig. ). The expression levels of Dmrt1, Dmrt2, Msl1 (male specific lethal-1), Msl2 (male specific lethal-2), Msl3 (male specific lethal-3), Fem1B, Fem1C, Sox3, Sox4, and Sox9 genes were approximately 76.1, 93.3, 14.5, 141, 51.5, 21.1, 186.3, 29.4, 77.9, and 4.8-fold higher, respectively, in spermary tissues than in ovarian tissues. The specificity of the real-time PCR products for the 10 novel transcripts was verified by sequencing.
Fig. 5. Expression analysis of 10 genes by real-time PCR.
Discussion
Functional classification and differential transcript analysis
We performed the first large-scale RNA sequencing of freshwater bivalves to provide insights into sex determination and support future studies. Only 21.31% of the 132,055 identified unigenes were successfully annotated. This result is consistent with the annotation of ESTs in the clams Sinonovacula constricta (20.7%) Citation26) and Meretrix meretrix (12.20%).Citation27) More annotation information was obtained using a domain search, but most of the functional sequence information remains unknown because of the limited genomic characterization of freshwater bivalves. However, some useful information can still be obtained from the annotated sequences. Many unigenes were matched with sequences from Saccoglossus kowalevskii (14.9%) and Strongylocentrotus purpuratus (7.4%), indicating that many of the physical processes in H. schlegelii share similarities with known pathways in these species. The COG analysis identified 9,399 unigenes, with the greatest number of annotated genes in four categories: predicted proteins (4231 candidates); proteins involved in DNA replication, recombination, and repair (1898); proteins involved in translation, ribosomal structure and biogenesis (1531); and proteins involved in transcription (1524). Many unique sequences were assigned to cell proliferation, reproduction, and reproductive processes, reflecting the biological processes responsible for oogenesis and spermatogenesis. A series of genes related to sex determination or reproduction were detected during these processes.
A total of 45,422 DEGs comprising 19,511 upregulated and 25,911 downregulated genes were detected in the spermary compared to the ovary. As expected, genes associated with oogenesis were highly upregulated in the ovary, whereas genes involved in spermatogenesis were highly upregulated in the spermary. Among the upregulated genes, five testis-specific and five sperm-specific expression genes were screened in the spermary, revealing significantly high expression of these genes in the sperm (Table ). Among the downregulated genes, nine genes associated with ovary development displayed higher expression in the ovary than in the spermary (Table ). After sex determination, the DEGs in the spermary and ovary participate in sexual differentiation, oogenesis, spermatogenesis, and fertilization in H. schlegelii.
Table 3. Genes differentially expressed in the spermary.
Table 4. Genes differentially expressed in the ovary.
Expression and regulation of genes related to sex determination—genes directly regulating male and female determination
Some sex-determination genes, such as WT1, Sf1, Sry, Sox9, GATA4, Dmrt1, and MIHCitation28) in males and Wnt4, RSpo1, Foxl2, and Dax1 in females,Citation29) have been studied in mammals. GATA4 and WT1 initiate the expression of Sry in the male sex-determination pathway.Citation30) Dmrt1 acts downstream of Sry,Citation3) leading to the expression of Sox9. The activated Sox9 further stimulates the expression of other key genes, including Fgf9 and PDGS, in testis differentiation and recruits Sertoli cells in the developing testis.Citation10) WT1, GATA4, Sry, Dmrt1, Dmrt2, Sox9, PDGS and many testis-specific expression genes were also screened in the sexual glands of H. schlegelii (Table ). Further expression analyses revealed that the expression levels of these genes were significantly higher in the spermary. Thus, male sex-determination mechanisms in H. schlegelii may be similar to those in mammals. There are two independent female sex-determination pathways for ovarian differentiation.Citation31) The Foxl2 gene has been identified as an important sex-determination gene. The ablation of Foxl2 in female mice induces the differentiation of ovarian cells into testicular cells.4) Significantly high expression of Foxl2 has also been observed in H. schlegelii ovaries, suggesting that Foxl2 may be an independent master gene responsible for ovarian development in H. schlegelii.Citation32) In another female sex-determination pathway, Rspo1 was identified as an ovarian determination gene that activates the Wnt signaling pathway during embryogenesis.Citation33,Citation34) β-catenin acts as a downstream effector of the Rspo1/Wnt signaling pathway. Intracellular β-catenin and TCF form a transcription complex that transactivates the target genes associated with ovarian differentiation. Similar to mammals, these female sex-determination genes exhibited significantly high levels of expression in the ovary of H. schlegelii, and the expression patterns of these genes were consistent with roles in ovary differentiation, as shown in mammals. These results suggest that the two female sex-determination pathways in mammals may also exist in H. schlegelii.
Table 5. Genes related to sex determination.
Expression of genes related to hermaphroditism
Some sex-determination genes, such as Msl, have been investigated in Drosophila.Citation35) Unequal numbers of X chromosomes were observed in females and males. However, the equal expression of X-linked genes between females and males is achieved through dosage compensation mechanisms.Citation36) Genetics studies have implicated three Msl genes, Msl1, Msl2, and Msl3 in dosage compensation;Citation37) these genes were also identified in the sexual glands of H. schlegelii. Real-time PCR analysis revealed that the expression levels of the three Msl genes were significantly higher in the sperm than in the ovary. Similar to Drosophila,Citation33) dosage compensation may occur in the spermary of H schlegelii via the twofold upregulation of gene expression on the single male X chromosome. Additional analyses are required to confirm this hypothesis.
The expression of sex genes may influence sex differentiation toward male, female, or hermaphrodite development. There are two sex phenotypes in C. elegans, hermaphrodite and male. Two important gene families involved in hermaphroditism, including Tra-1, Tra-2α, Tra-2β, Fem1A, Fem1B, and Fem1C, have been investigated in C. elegans.Citation38) In the hermaphrodite regulation mechanism, Tra-2 regulates the activity of Tra-1 through the suppression of Fem proteins. When activated Tra-1 inhibits the expression of male development genes, a hermaphroditic individual developsCitation15). These two gene families were screened in the sexual glands of H. schlegelii. The transcript analysis revealed that the expression levels of Tra-1, Tra-2α, and Tra-2β were higher in the ovary than in the spermary. However, the expression levels of Fem1A, Fem1B, and Fem1C were significantly higher in the sperm than in the ovary. Thus, these six genes may directly regulate the hermaphroditic phenomenon. Further transcriptomic analysis of the sex determination genes may provide additional insights into the mechanisms underlying hermaphroditism in freshwater bivalves.
Conclusion
In the present study, we present the first use of Illumina sequencing technology in freshwater bivalves without prior genome annotation. The large-scale transcriptome of H. schlegelii was successfully sequenced using de novo assembly. These findings substantially contribute to existing sequence resources and accelerate the progress of research on sex determination and differentiation in freshwater bivalves. These sequence data are also a useful tool for future studies involving gene expression experiments, comparative transcriptomics, genomics, and metabolomics in freshwater bivalves.
Author contributions
Conceived the experiments: Yijiang Hong. Designed and Performed the experiments: Jianwu Shi, Kou Peng, Junqing Sheng. Analyzed the data: Jianwu Shi. Contributed reagents/materials/analysis tools: Junhua wang. Wrote the paper: Jianwu Shi, Yijiang Hong.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921–3930.10.1242/dev.048983
- Jakob S, Lovell-Badge R. Sex determination and the control of Sox9 expression in mammals. FEBS J. 2011;278:1002–1009.10.1111/j.1742-4658.2011.08029.x
- Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends in Genet. 2012;28:175–184.10.1016/j.tig.2012.02.002
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, Riethmacher D, Schütz G, Cooney AJ, Lovell-Badge R, Treier M. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142.
- Chassot AA, Gregoire EP, Lavery R, Taketo MM, de Rooij DG, Adams IR, Chaboissier MC. RSPO1/β-catenin signaling pathway regulates oogonia differentiation and entry into meiosis in the mouse fetal ovary. PLoS One. 2011;6:e25641.10.1371/journal.pone.0025641
- Lu H, Huang X, Zhang L, Guo Y, Cheng H, Zhou R. Multiple alternative splicing of mouse Dmrt1 during gonadal differentiation. chem. Biophys. Res. Commun. 2007;352:630–634.10.1016/j.bbrc.2006.11.066
- Chue J, Smith CA. Sex determination and sexual differentiation in the avian model. FEBS J. 2011;278:1027–1034.10.1111/j.1742-4658.2011.08032.x
- Aoyama S, Shibata K, Tokunaga S, Takase M, Matsui K, Nakamura M. Expression of Dmrt1 protein in developing and in sex-reversed gonads of amphibians. Cytogenet. Genome Res. 2003;101:295–301.10.1159/000074352
- Herpin A, Schartl M. Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J. 2011;278:1010–1019.10.1111/j.1742-4658.2011.08030.x
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin d synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 2007;282:10553–10560.10.1074/jbc.M609578200
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends in Genet. 2009;25:19–29.10.1016/j.tig.2008.10.008
- Jiang T, Hou CC, She ZY, Yang WX. The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol. Biol. Rep. 2013;40:2187–2194.10.1007/s11033-012-2279-3
- Hempel LU, Oliver B. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC. Dev. Biol. 2007;7:113.10.1186/1471-213X-7-113
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480.
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell. 2007;13:127–139.10.1016/j.devcel.2007.05.008
- Haag ES, Wang S, Kimble J. Rapid coevolution of the Nematode sex-determining genes fem-3 and tra-2. Current Biol. 2002;12:2035–2041.10.1016/S0960-9822(02)01333-7
- Yu Y, Hong YJ, Qiu QJ, Wang JH, Xu MX, Li YJ. Embryonic development and breading-season gonad in Hyriopsis schlegeli. Chn. J. Zool. 2008;43:102–107.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, Chen ZH, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652.
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676.10.1093/bioinformatics/bti610
- Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297.
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 2008;5:621–628.10.1038/nmeth.1226
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome. Res. 1997;7:986–995.
- Shi JW, Peng K, Sheng JQ, Wang JH, Yi WJ, Wu HJ, Gu Q, Hong YJ. A specific genomic organization and a novel promoter sequence for both ZP2 and ZP3 gene expressions in the Pingxiang red transparent crucian carp, Carassius auratus var. pingxiangnensis. Comp. Biochem. Physiol. D: Genomics Proteomics. 2013;8:275–282.
- Shi JW, Sheng JQ, Peng K, Wang JH, Yi WJ, Wu HJ, Gu Q, Hong YJ. Expression pattern of the zona pellucida 3 (ZP3) gene during ovarian development and the location of ZP3 protein in oocytes in a natural, wild triploid crucian carp mutant, Carassius auratus var. Pingxiangnensis. Genet. Mol. Res. 2013;12:5640–5650.10.4238/2013.November.18.13
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s C T difference” formula. J. Mol. Med. 2006;84:901–910.10.1007/s00109-006-0097-6
- Feng B, Dong L, Niu D, Meng S, Zhang B, Liu D, Hu S, Li J. Identification of immune genes of the Agamaki Clam (Sinonovacula constricta) by sequencing and bioinformatic analysis of ESTs. Mar. Biotechnol. 2010;12:282–291.10.1007/s10126-009-9216-z
- Huan P, Wang HX, Liu BZ. Transcriptomic analysis of the clam Meretrix meretrix on different larval stages. Mar. Biotechnol. 2012;14:69–78.10.1007/s10126-011-9389-0
- Ferguson-Smith M. The evolution of sex chromosomes and sex determination in vertebrates and the key role of DMRT1. Sex. Dev. 2007;1:2–11.10.1159/000096234
- Wilhelm D. R-spondin1-discovery of the long-missing, mammalian female-determining gene? BioEssays. 2007;29:314–318.10.1002/(ISSN)1521-1878
- Miyamoto Y, Taniguchi H, Hamel F, Silversides DW, Viger RS. A GATA4/WT1 cooperation regulates transcription of genes required for mammalian sex determination and differentiation. BMC Mol. Biol. 2008;9:44.10.1186/1471-2199-9-44
- Kocer A, Pinheiro I, Pannetier M, Renault L, Parma P, Radi O, Kim KA, Camerino G, Pailhoux E. R-spondin1 and FOXL2 act into two distinct cellular types during goat ovarian differentiation. BMC Dev. Biol. 2008;8:36.10.1186/1471-213X-8-36
- Matsumoto Y, Crews D. Molecular mechanisms of temperature-dependent sex determination in the context of ecological developmental biology. Mol. Cell. Endocrinol. 2012;354:103–110.10.1016/j.mce.2011.10.012
- Lau YFC, Li YM. The human and mouse sex-determining SRY genes repress the Rspol/β-catenin signaling. J. Genet. Genomics. 2009;36:193–202.10.1016/S1673-8527(08)60107-1
- Tomaselli S, Megiorni F, Lin L, Mazzilli MC, Gerrelli D, Majore S, Grammatico P, Achermann JC. Human RSPO1/R-spondin1 is expressed during early ovary development and augments β-catenin signaling. PLoS ONE. 2011;6:e16366.10.1371/journal.pone.0016366
- Baker BS, Gorman M, Marin I. Dosage compensation in Drosophila. Annu. Rev. Genet. 1994;28:491–521.10.1146/annurev.ge.28.120194.002423
- Marín I, Siegal ML, Baker BS. The evolution of dosage-compensation mechanisms. BioEssays. 2000;22:1106–1114.10.1002/1521-1878(200012)22:12<>1.0.CO;2-8
- Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 2006;20:871–883.10.1101/gad.377506
- Madl JE, Herman RK. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 1979;93:393–402.