Abstract
Cells respond to the environment and alter gene expression. Recent studies have revealed the social aspects of bacterial life, such as biofilm formation. Biofilm formation is largely affected by the environment, and the mechanisms by which the gene expression of individual cells affects biofilm development have attracted interest. Environmental factors determine the cell’s decision to form or leave a biofilm. In addition, the biofilm structure largely depends on the environment, implying that biofilms are shaped to adapt to local conditions. Second messengers such as cAMP and c-di-GMP are key factors that link environmental factors with gene regulation. Cell-to-cell communication is also an important factor in shaping the biofilm. In this short review, we will introduce the basics of biofilm formation and further discuss environmental factors that shape biofilm formation. Finally, the state-of-the-art tools that allow us investigate biofilms under various conditions are discussed.
Graphical abstract
Environmental factors affect biofilm formation throughout their development.
Key words:
Biofilms are structured aggregates of bacterial cells that are embedded in self-produced extracellular polymeric substances (EPS).Citation1,2) Biofilms may have been part of the earliest life on Earth;Citation3) and most bacteria are now thought to form biofilms in nature.Citation4,5) Biofilm-forming cells usually differ from their planktonic counterparts and exhibit differences in gene expression.Citation6,7) One of the characteristics of biofilms is that the interior environment is heterogeneous, and thus gene expression profiles that are specific to cell location in the biofilm are observed.Citation8) The multicellularity of biofilms suggests that these cells develop a social network with division of labor. The emerging field of microbiology that studies the social aspects of bacterial life, including biofilm formation, is now recognized as “sociomicrobiology”.Citation9)
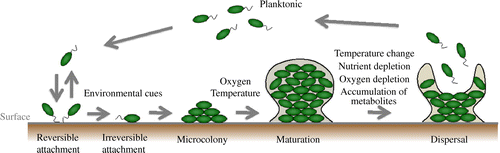
Biofilm formation is generally established through several steps. First, planktonic cells attach to the surface in a process known as surface attachment. After surface attachment, biofilms form a structured architecture with the assistance of EPS in the maturation stage. After maturation, the cell leaves the biofilm in the dispersal stage. Each stage is affected by several environmental factors and is a highly regulated process (Fig. ).Citation10,11)
Fig. 1. Environmental factors that shape biofilm formations.
Notes: The image shows biofilm formation and selected environmental factors that affect each stage of biofilm formation. The environmental cues control the concentration of second messengers such as cAMP and c-di-GMP. These second messengers control biofilm-related factors such as cell appendages, surface proteins, EPS, and cell motility. QS is also involved in shaping the biofilm by controlling the above biofilm-related factors.
1. Surface attachment
Surface attachment is one of the most important processes because it represents a turning point from planktonic life to the biofilm mode. Surface attachment has been studied in detail in Pseudomonas aeruginosa which has been employed as a model organism for biofilm formation. First, bacterial cells attach to the surface reversibly at the pole of the cells, which is called reversible attachment.Citation11,12) Cell appendages such as flagella, pili, and fimbriae are involved in the reversible attachment. At this stage, the bacteria can commit to the biofilm lifestyle or leave the surface and return to the planktonic lifestyle. After reversible attachment, cells undergo irreversible attachment.Citation13) At this stage, surface proteins such as SadB or LapA and EPS assist the adhesion between the cell and surface.Citation14,15) During the transition to irreversible attachment, an intracellular second messenger, bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP), is involved. c-di-GMP is produced by many bacteria and regulates EPS production and motility in opposing directions. In addition, Ono et al. have reported that another second messenger, cAMP, is involved in the transition from reversible to irreversible attachment.Citation16) c-di-GMP and cAMP concentrations are controlled by various environmental conditions, such as carbon and oxygen, and thus the regulation of surface attachment by these second messengers reflects environmental conditions.Citation17) It was recently demonstrated that differences in attachment ability play a role in speciation, emphasizing the importance of surface attachment.Citation18)
2. Biofilm maturation
After irreversible attachment, bacterial cells start to develop microcolonies by assembling previously attached cells and undergoing cell division.Citation19) These microcolonies grow via cell proliferation and produce EPS. In most mature biofilms, EPS represents more than 90% of the dry mass.Citation1) EPS components include polysaccharides, nucleic acids, proteins, lipids, and other biopolymers. In the biofilm, EPS is responsible for adhesion to surfaces, scaffolding cells together, and maintaining the three-dimensional architecture of the biofilm. Furthermore, the EPS surrounding the biofilm protects the bacterial cells against various stresses such as antimicrobials, host immune systems, oxidation and metallic cations.Citation1) Inside the biofilm, EPS retains quorum sensing (QS) signaling molecules, extracellular enzymes, and metabolic products. Therefore, EPS supports cell–cell communication and degrading substances.Citation1,20)
Advances in analytical methods have revealed that a biofilm is not a simple aggregation of bacterial cells and EPS but has a complex architecture.Citation10) P. aeruginosa and Bacillus subtilis form biofilms interspersed with open water channels.Citation10,21,22) These channels may facilitate the exchange of nutrients and waste products by infusing fluid into the biofilm.Citation10) Interestingly, several studies have demonstrated that biofilm structure changes dynamically depending on the environmental conditions. For example, P. aeruginosa, the model biofilm-forming bacterium, forms a mushroom-shaped structure with channels between macrocolonies consisting of rod-shaped cells under oxic conditions. By contrast, P. aeruginosa forms a three-dimensional mesh-like structure with channels or spaces between macrocolonies consisting of elongated filamentous cells under anoxic conditions.Citation23) This morphological change in the biofilm presumably alters the diffusibility of substances and enables metabolic adaptation under oxic or anoxic conditions by optimizing nutrient and waste product exchange. However, the effect of anaerobiosis varies among strains, suggesting complicated regulation of biofilm formation.Citation24) In addition, it was recently demonstrated that a respiratory enzyme affects biofilm formation, indicating roles other than energy conservation in the formation of a multicellular community under certain conditions.Citation25) In Clostridium perfringens, a gram-positive, spore-forming, anaerobic pathogen, temperature changes drastically affect biofilm morphology, including thickness and cell density, reflecting the temperature-dependent regulation of EPS production (Fig. ).Citation26) Temperature is thus an important signal of the environment outside or inside the mammalian host, ensuring that temperature-regulated biofilm formation is induced appropriately for infection.Citation26) Biofilm structure is therefore not simple and uniform but dynamically changes depending on environmental conditions. Biofilm formation seems to be a preparation for unstable environmental conditions. The notion that biofilm formation is an “insurance effect” of cells against changing environments is supported by the more frequent isolation of variants from biofilms than from planktonic cultures.Citation27)
Fig. 2. The images show different biofilm structures of C. perfringens under different temperatures.
Notes: Left panels 37 °C. Right panels 25 °C. (A) Pellicle biofilm formation at 25 °C. (B) Crystal violet staining of surface-attached biofilms. (C and D) Confocal reflection microscopy images (scale bar, 20 μm). (E) Scanning electron microscopy images of the biofilms (scale bar, 2 μm). Image is reprinted from ref 26. Copyright © 2014, American Society for Microbiology [Journal of bacteriology, 196, 2014, 1540–1550, doi:10.1128/JB.01444-13].
![Fig. 2. The images show different biofilm structures of C. perfringens under different temperatures.Notes: Left panels 37 °C. Right panels 25 °C. (A) Pellicle biofilm formation at 25 °C. (B) Crystal violet staining of surface-attached biofilms. (C and D) Confocal reflection microscopy images (scale bar, 20 μm). (E) Scanning electron microscopy images of the biofilms (scale bar, 2 μm). Image is reprinted from ref 26. Copyright © 2014, American Society for Microbiology [Journal of bacteriology, 196, 2014, 1540–1550, doi:10.1128/JB.01444-13].](/cms/asset/6a6e7fec-c090-4a23-85f8-bf8b8c10102f/tbbb_a_1058701_f0002_b.gif)
3. Biofilm dispersion
From mature biofilm, some bacterial cells transfer to planktonic growth. These dispersed cells explore other niches and attach again to a new surface. Thus, dispersal is not only the final stage of the biofilm lifecycle but also the start of another lifecycle. There is “active dispersal”, which depends on cell motility or degradation of EPS, and “passive dispersal”, which depends on physical factors such as shearing force under liquid flow conditions.Citation10,28) Active dispersal is triggered by changes in environmental conditions such as temperature change, starvation, oxygen deficiency, and metabolite accumulation.Citation28) In active dispersal, genes involved in cell motility, such as flagella synthesis or chemotaxis and in EPS degradation, such as dispersin secretion, are upregulated, while genes involved in EPS production, such as polysaccharide synthesis, and in attachment, such as fimbriae synthesis, are downregulated.Citation28) The intracellular second messenger c-di-GMP is closely involved in this gene regulation. Intracellular c-di-GMP levels are dynamically controlled by environmental conditions. In contrast to the attachment stage, a decrease in intracellular c-di-GMP levels promotes the conversion from the planktonic to biofilm mode of life by regulating genes involved in dispersal. By contrast, an increase in intracellular c-di-GMP levels promotes conversion to the biofilm mode of life. Thus, c-di-GMP plays a central role as a switch between the planktonic and biofilm modes of life.Citation28,29)
4. Cell-to-cell communication in bacteria
Cell-to-cell communication plays a role in the organization and differentiation of the cell in biofilms depending on conditions. Bacteria communicate using various chemical signal compounds. One type of communication used to recognize the population density of the same species is QS, which plays an important role in bacterial cell-to-cell communication.Citation30) Bacteria produce signal compounds, which accumulate with increasing cell density. After the concentration of the signal reaches a threshold level, it is recognized by receptor(s) located in the cytoplasm or cell membrane and activates gene expression involved in signal production. This feed-forward auto regulation loop of QS genes promotes synchronization of the cell community in terms of the QS response.Citation31) QS synchronously regulates specific genes that facilitate various group activities such as bioluminescence and the production of extracellular enzymes and toxins. Pathogenic bacteria do not express virulence factors at low cell density and thus escape from the host immune response. However, when grown at high cell density under optimal growth conditions, pathogenic bacteria collectively produce toxins. The QS system enforces synchrony in group behaviors and presumably optimizes group activities until the cost balances the benefit. Bacteria use diverse signal molecules and different types of QS systems. Gram-negative bacteria produce acyl-homoserine lactone and derivatives of S-adenosyl methionine (SAM) such as AI-2. Gram-positive bacteria use peptides called autoinducing peptides (AIPs) as signaling molecules, and actinomycete produces A-factor.Citation32) The definition of signaling molecules for cell-to-cell communication such as QS varies among different researchers, and the term signals is broadly used because QS signal molecules could also act as biological cues that affect individual phenotypes outside QS systems. However, the generally accepted definition is that signals benefit the sender and not the receiver and that signal production coevolves with its response.Citation33) The QS system is involved not only in virulence factor production but also primary metabolism such as respirationCitation34,35) and biofilm formation, and the effective utilization of signaling molecules is expected to permit control methods for the microbial community . Bacterial MVs were recently implicated in the delivery of signaling molecules.Citation36) Several pathways of MV biogenesis have been proposed, and MV quantity and quality likely depends on environmental conditions.Citation36,37)
5. Relationship between biofilm formation and cell-to-cell communication
Cells are densely packed in biofilms, enabling the high accumulation of signaling molecules as well as metabolites and secretion products. Subsequent to the report by Davies et al. (1998) of the relationship between biofilm formation and cell-to-cell communication in P. aeruginosa,Citation38) biofilm regulation by QS systems has been investigated in various bacteria. The agr QS mutant of Staphylococcus aureus increases biofilm formation under specific conditions, and the addition of Agr peptide signals to the culture induces biofilm dispersal.Citation39) The Vibrio cholera QS system, which uses signals derived from SAM, inhibits biofilm formation by inducing the expression of the master transcriptional regulator HapR.Citation40) Bacterial biofilms are capable of forming elaborate conformations, and QS signaling has also been suggested to be involved in structural determination. P. aeruginosa QS systems regulate swarming motility and the release of extracellular DNAs, thereby influencing biofilm structures.Citation41) In addition, approximately 30% of biofilm EPS proteins are derived from MVs,Citation42) suggesting that MVs are abundant in biofilms and may also play a role in the delivery of signal compounds. These important relationships between cell-to-cell communication and biofilm formation are profoundly influenced by the experimental conditions, such as medium, and many bacteria are capable of forming structural biofilms without cell-to-cell communication. Thus, the contribution of cell-to-cell communication to biofilm structures may be conditional and requires further study. The lack of clarity is due to the limited analytical methods available for biofilms, which include microscopy and antibiotic tolerance tests. The physiological state of biofilm cells is poorly understood. Therefore, to clarify the relationship between biofilm formation and cell-to-cell communication, new innovative techniques are needed that can observe biofilm architecture continuously and dynamically, and simultaneously analyze physiological states.
6. Development of biofilm analysis technologies
As indicated above, previous studies have revealed that most bacteria form functional structured biofilms whose formation is regulated by various endogenous and exogenous factors. These findings were made possible by the development of analysis technologies. Therefore, the development and improvement of biofilm analysis technologies are critical to biofilm studies.
7. Basic technologies for analyzing biofilms
At the dawn of biofilm research, scanning electron microscopy (SEM) was frequently used to observe the detailed structure of biofilms.Citation43,44) Although SEM revealed the fine structure of biofilms at the nanometer scale, SEM sample preparation requires invasive treatments such as fixing, dehydration, and drying. Thus, live biofilms cannot be observed by SEM. Fluorescent imaging methods using confocal laser scanning microscopy (CLSM) were subsequently established for real-time imaging of biofilms.Citation45) Biofilm observation methods using CLSM and fluorescent protein expression systems have become mainstream in biofilm studies. Moreover, the combination of continuous culture systems and CLSM has been broadly used to observe the biofilm formation process.Citation46,47) These visualization technologies and methods have allowed raw biofilms to be observed sequentially to reveal three-dimensional structures and the formation process. However, the application range of fluorescent microscopy is very restricted because of the requirement for genetic transformation and moderate culture conditions for normal fluorescent protein function.Citation45)
Flow-cell systems combined with CLSM are generally used to observe the detailed three-dimensional structure during biofilm development. However, conventional techniques to observe biofilms, including the flow-cell system, largely depend on fluorescence. In this case, cells must be stained with fluorescent dyes or tagged with fluorescent proteins. The reflection microscopy-based method continuous-optimizing reflection microscopy (COCRM) was developed to address this technical limitation. The key difference is that COCRM uses reflected light as the signal, rather than fluorescence in CLSM.Citation48) Thus, COCRM does not depend on fluorescence and permits three-dimensional visualization of biofilms without genetic transformation or fluorescent probing.Moreover, COCRM uniquely enables non-destructive visualization of the biofilm, including its attached substratum. Recently, the sequential visualization of an oral complex biofilm with its attached substratum and biofilm-related dental caries was reported.Citation49) Furthermore, a combination of COCRM and fluorescent staining has been used to visualize substance transport into the biofilm.Citation50)
The more challenging issue in biofilm research is to measure the physiological state of the cells. To this end, metabolites released from the cells are measured. Water-soluble metabolites along with the three-dimensional structures of biofilms have been analyzed.Citation51) Gaseous metabolites that are produced as a result of respiration in biofilms have also been analyzed using a flow culture system named “Airtight Flow reactor for nondestructive Gaseous metabolite Analysis and Structure visualization” (AFGAS).Citation23) The AFGAS method permits the simultaneous and sequential analysis of biofilm structure, aqueous metabolites, and gaseous metabolites. The relationship between anaerobic respiration and biofilm structure in P. aeruginosa has been examined using the AFGAS method.Citation23) As shown above, sequential analysis of biofilms has become possible via the development of visualization technologies. However, sequential and simultaneous analysis requires a large flow system and is not suitable for multiple samples and multi-condition experiments. Microfluidic device technology can provide ground-breaking experimental solutions in biofilm research. Microfluidic device technology permits the fabrication of a variety of miniature experimental device(s), such as microculture chambers, microchannels, microvalves, or microsensors, on a small (typically less than one square inch) silicone chip. Microfluidic devices offer unprecedented flexibility in configuration and precise control of the internal environment, enabling new approaches for questions that were previously difficult to address. Microenvironments such as micropatches of nutrient or shear force can be developed in the devices, making them particularly suitable for analyzing bacterial ecology in response to the environment.Citation18,52) Moreover, microfluidic devices can be easily designed to integrate microsensors that permit the rapid detection of cells.Citation53) Recently, a microfluidic device was recently developed that maintains cells in a small culture chamber in which an on-chip microsensor continuously monitors ammonia consumption and a transparent silicon material allows continuous cell visualization by COCRM.Citation54,55) Precise analysis of the relationship between temporal shifts in cell morphology and ammonia metabolism is possible due to the unique characteristics of microfluidics. Microfluidic technologies are progressively becoming a tool of choice for a wide range of microbial studies.Citation18,56,57)
8. Future perspectives
It is increasingly evident that bacterial societies are more complex than once thought. Gene expression in bacterial communities is heterogenous,Citation58–60) and subpopulations that exhibit different phenotypes, such as persister cells,Citation61) emerge. While most laboratory biofilm studies are conducted under static conditions, natural conditions fluctuate. How this heterogeneous gene expression contributes to the flexibility and plasticity of biofilms in fluctuating environments is poorly understood, and its elucidation would not only deepen our knowledge but also permit the development of new ways in treating bacteria. The development of techniques such as single-cell analysis will enable the investigation of more complex communities, which may allow us explore the multicellularity of this unicellular organism.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This study was supported in part through funding from Grant-in-aid for Scientific Research to M.T., N.O. and N. N. from The Ministry of Education, Culture, Sports, and Technology of Japan. T. K. was funded by JSPS. This research was also funded through the Japan Science and Technology Agency, CREST, and ALCA.
References
- Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633.
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79.10.1146/annurev.micro.54.1.49
- Westall F, de Wit MJ, Dann J, van der Gaast S, de Ronde CEJ, Gerneke D. Early Archean fossil bacteria and biofilms in hydrothermally-influenced sediments from the Barberton greenstone belt, South Africa. Precambrian Res. 2001;106:93–116.10.1016/S0301-9268(00)00127-3
- Lerchner J, Wolf A, Buchholz F, et al. Miniaturized calorimetry—A new method for real-time biofilm activity analysis. J. Microbiol. Methods. 2008;74:74–81.10.1016/j.mimet.2008.04.004
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002;56:187–209.10.1146/annurev.micro.56.012302.160705
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193.10.1128/CMR.15.2.167-193.2002
- Nickel JC, Ruseska I, Wright JB, Costerton JW. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 1985;27:619–624.10.1128/AAC.27.4.619
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953.10.1101/gad.1645008
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33.10.1016/j.tim.2004.11.007
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108.10.1038/nrmicro821
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998;28:449–461.10.1046/j.1365-2958.1998.00797.x
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304.10.1046/j.1365-2958.1998.01062.x
- Petrova OE, Sauer K. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 2012;194:2413–2425.10.1128/JB.00003-12
- Caiazza NC, O’Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 2004;186:4476–4485.10.1128/JB.186.14.4476-4485.2004
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918.10.1046/j.1365-2958.2003.03615.x
- Ono K, Oka R, Toyofuku M, et al. cAMP signaling affects irreversible attachment during biofilm formation by Pseudomonas aeruginosa PAO1. Microbes Environ. 2014;29:104–106.10.1264/jsme2.ME13151
- McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 2012;10:27–38.
- Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5622–5627.10.1073/pnas.1318943111
- Zhao K, Tseng B, Beckerman B, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–391.10.1038/nature12155
- Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 2014;24:50–55.10.1016/j.cub.2013.10.030
- Stoodley P, Debeer D, Lewandowski Z. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 1994;60:2711–2716.
- Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 2013;110:848–852.10.1073/pnas.1216376110
- Yawata Y, Nomura N, Uchiyama H. Development of a novel biofilm continuous culture method for simultaneous assessment of architecture and gaseous metabolite production. Appl. Environ. Microbiol. 2008;74:5429–5435.10.1128/AEM.00801-08
- Fang H, Toyofuku M, Kiyokawa T, Ichihashi A, Tateda K, Nomura N. The impact of anaerobiosis on strain-dependent phenotypic variations in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 2013;77:1747–1752.10.1271/bbb.130309
- Hamada M, Toyofuku M, Miyano T, Nomura N. cbb3-type cytochrome c oxidases, aerobic respiratory enzymes, impact the anaerobic life of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014;196:3881–3889.10.1128/JB.01978-14
- Obana N, Nakamura K, Nomura N. A sporulation factor is involved in the morphological change of Clostridium perfringens biofilms in response to temperature. J. Bacteriol. 2014;196:1540–1550.10.1128/JB.01444-13
- Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12503–12508.10.1073/pnas.0801499105
- McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2012;10:39–50.
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52.10.1128/MMBR.00043-12
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275.
- Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:a012427.
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246.10.1016/j.cell.2006.04.001
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 2013;67:43–63.10.1146/annurev-micro-092412-155635
- Toyofuku M, Nomura N, Fujii T, et al. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2007;189:4969–4972.10.1128/JB.00289-07
- Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 2008;190:7947–7956.10.1128/JB.00968-08
- Tashiro Y, Ichikawa S, Shimizu M, et al. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010;76:3732–3739.10.1128/AEM.02794-09
- Tashiro Y, Uchiyama H, Nomura N. Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ. Microbiol. 2012;14:1349–1662.10.1111/j.1462-2920.2011.02632.x
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298.10.1126/science.280.5361.295
- Boles BR, Horswill AR. agr-Mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052.
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholera. Dev. Cell. 2003;5:647–656.10.1016/S1534-5807(03)00295-8
- de Kievit TR. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009;11:279–288.10.1111/emi.2009.11.issue-2
- Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 2012;11:4906–4915.10.1021/pr300395j
- Korber DR, Lawrence JR, Sutton B, Caldwell DE. The effect of laminar flow on the kinetics of surface recolonization by mott and mot- Pseudomonas fluorescens. Microb. Ecol. 1989;18:1–19.10.1007/BF02011692
- Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE. Optical sectioning of microbial biofilms. J. Bacteriol. 1991;173:6558–6567.
- Palmer RJ, Sternberg C. Modern microscopy in biofilm research: confocal microscopy and other approaches. Curr. Opin. Biotechnol. 1999;10:263–268.10.1016/S0958-1669(99)80046-9
- Korber DR, Wolfaardt GM, Brözel V, MacDonald R, Niepel T. Reporter systems for microscopic analysis of microbial biofilms. Methods Enzymol. 1999;310:3–20.
- McLean RJ, Bates CCL, Barnes MB, McGowin CL, Aron GM. Methods of studying biofilms. In: Ghannoum M, O’Tool GA, editors. Microbial biofilms. Washington, DC: ASM Press; 2004. p. 379–413.
- Yawata Y, Toda K, Setoyama E, et al. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J. Biosci. Bioeng. 2010;110:377–380.10.1016/j.jbiosc.2010.04.002
- Inaba T, Ichihara T, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Three- dimensional visualization of mixed species biofilm formation together with its substratum. Microbiol. Immunol. 2013;57:589–593.10.1111/mim.v57.8
- Yawata Y, Uchiyama H, Nomura N. Visualizing the effects of biofilm structures on the influx of fluorescent material using combined confocal reflection and fluorescent microscopy. Microbes Environ. 2010;25:49–52.10.1264/jsme2.ME09169
- Zhu Y, Weiss EC, Otto M, Fey PD, Smeltzer MS, Somerville GA. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 2007;75:4219–4226.10.1128/IAI.00509-07
- Rusconi R, Garren M, Stocker R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 2014;43:65–91.10.1146/annurev-biophys-051013-022916
- Richter L, Stepper C, Mak A, et al. Development of a microfluidic biochip for online monitoring of fungal biofilm dynamics. Lab Chip. 2007;7:1723–1731.10.1039/b708236c
- Toda K, Yawata Y, Setoyama E, Fukuda J, Nomura N, Suzuki H. Continuous monitoring of ammonia removal activity and observation of morphology of microbial complexes in a microdevice. Appl. Environ. Microbiol. 2011;77:4253–4255.10.1128/AEM.01246-10
- Yawata Y, Toda K, Setoyama E, et al. Bacterial growth monitoring in a microfluidic device by confocal reflection microscopy. J. Biosci. Bioeng. 2010;110:130–133.10.1016/j.jbiosc.2010.01.009
- Kim J, Park HD, Chung S. Microfluidic approaches to bacterial biofilm formation. Molecules. 2012;17:9818–9834.10.3390/molecules17089818
- Wessel AK, Hmelo L, Parsek MR, Whiteley M. Going local: technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 2013;11:337–348.10.1038/nrmicro3010
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186.10.1126/science.1070919
- Williamson KS, Richards LA, Perez-Osorio AC, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 2012;194:2062–2073.10.1128/JB.00022-12
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953.10.1101/gad.1645008
- Lewis K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007;5:48–56.10.1038/nrmicro1557
