Abstract
Rice α-fucosidase (α-fucosidase Os, 58 kDa) that is active for α1-4 fucosyl linkage in Lewis a unit of plant N-glycans was purified to homogeneity. α-fucosidase Os showed activity against α1-3 fucosyl linkage in Lacto-N-fucopentaose III but not α1-3 fucosyl linkage in the core of plant N-glycans. The N-terminal sequence of α-fucosidase Os was identified as A-A-P-T-P-P-P-L-, and this sequence was found in the amino acid sequence of the putative rice α-fucosidase 1 (Os04g0560400).
Structural features of plant N-glycans are the occurrence of β1,2-xylosyl (Xyl) and α1,3-fucosyl (Fuc) residues linked to the trimannosyl core structure (Man3GlcNAc2). In addition to the α1,3-Fuc residue, α1,4-Fuc residue(s) is/are found in the Lewis a epitope occurring often on secreted type plant N-glycoproteins. Plant α-fucosidase (α-fucosidase), which hydrolyzes α1,3- and α1,4-linkages of Fuc to GlcNAc in Lewis-type glycans, was purified from almond meal (designated as almond α-fucosidase I) and the detail substrate specificity was determined.Citation1–4) However, any plant α-fucosidase being active against the α1,3-Fuc linked to the innermost GlcNAc residue of the plant complex type (PCT) N-glycans has not been characterized so far. Zeleny et al. have cloned an Arabidopsis thaliana α-fucosidase based on the partial amino acid sequences of the almond α-fucosidase I and confirmed the α-fucosidase activity of the recombinant protein.Citation5) It was found that the recombinant α-fucosidase was active against the α1,4-fucosyl residue in the Lewis a epitope and the α1,3-fucosyl residue in the lacto-N-fucopentaose III (LNFP III) but not the α1,3-fucosyl residue in one of fucosylated small N-glycans (Man3Fuc1GlcNAc2). In this study, therefore, we have tried to purify and characterize α-fucosidase from rice culture cells that hydrolyze the α1,3-fucosyl linkage in the PCT N-glycans.
Rice k-1 cell line used in this study was established from Oryza sativa L. cv. Nipponbare by Professor K. Kasamo (Research Institute for Bioresources, Okayama University). Crude enzyme was extracted from the rice culture cells (170 g, wet weight) suspended in 20 mM Tris-HCl buffer (pH 8.0) by sonication. From the crude extract, rice α-fucosidase was purified by a combination of gel-filtration (Sephadex G-75), ion-exchange chromatography (Q-Sepharose and Shodex QAE column), hydrophobic interaction chromatography (Shodex Phenyl column), and gel filtration (TSK-Gel G3000 SWXL). Through all of the purification steps, the activity of α-fucosidase was measured using a pyridylaminated plant complex type N-glycans bearing Lewis a epitope (Gal2Fuc2GlcNAc2Man3Xyl1Fuc1GlcNAc-PA, Gal2Fuc2GN2M3FX)Citation6) as a substrate, since we were not able to detect the α-fucosidase activity when pNP-α-fucoside was used as a substrate. The enzyme solution (10–20 µL) was added to 2 µL of Gal2Fuc2GN2M3FX (7.2 pmol/µL) in 0.1 M Na-acetate buffer (pH 5.0), and the reaction mixture was incubated at 37 °C for 12 h. The defucosylated substrates were analyzed by size fractionation HPLC (SF-HPLC) as shown in Fig. , and the amount of digested substrate (the reduced amount of original substrate) was quantified by SF-HPLC. One unit of enzyme activity was defined as the amount of enzyme digesting 1 nmol of the substrate per min at 37 °C. The products were analyzed with a Jasco 880-PU HPLC apparatus with a Jasco Intelligent spectrofluorometer (Jasco, Tokyo) and a Shodex Asahipak NH2P-50 column (4.6 × 250 mm, Showa Denko, Tokyo) at a flow rate of 0.7 mL/min using two-solvent system (80% acetonitrile/water and 20% acetonitrile/water) as described in our previous paper.Citation7) PA-sugar chains were detected with a Jasco FP-920 Intelligent Fluorescence detector (excitation 310 nm, emission 380 nm).Citation7) When the pyridylaminated lacto-N-fucopentaose III (LNFP III, Takara Bio Inc., Japan) was used as a substrate, the products obtained were analyzed with a Cosmosil 5C18-AR column (6.0 × 250 mm) at a flow rate of 1.0 mL/min using two-solvent system (0.02% TFA/water and 20% acetonitrile/water) described in our previous report.Citation8)
Fig. 1. Substrate specificity of rice α-fucosidase (α-fucosidase Os).
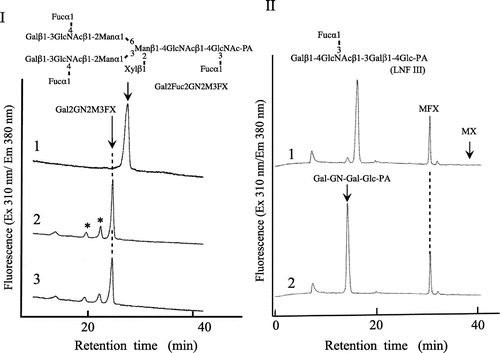
Rice α-fucosidase (α-fucosidase Os, α-fucosidase from Oriza sativa) was purified about 100-fold to homogeneity (total units, 0.21 mU; specific activity, 16.9 mU/mg), as shown in Fig. . Although the purification fold was about 480 at the penultimate purification step (Phenyl-HPLC), the purification fold decreased to about 100 after the final purification step (TSK-Gel G3000SWXL), suggesting that some unknown factor(s) would be necessary for the full activity. The molecular mass of the purified α-fucosidase Os was estimated to be about 58 kDa on SDS-PAGE under reducing and non-reducing condition (Fig. ). The maximum level of activity was obtained at pH 5.5 and the optimum temperature was 50 °C at pH 5.0.
Fig. 2. SDS-PAGE of purified α-fucosidase Os and deduced amino acid sequence of putative rice α-fucosidase 1.
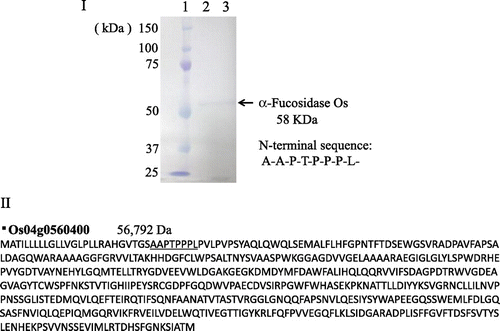
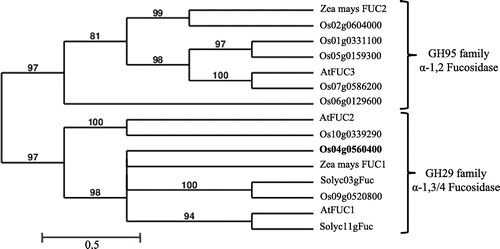
Fig. 3. Phylogenetic tree of plant α-fucosidases.
As well as Arabidopsis α-fucosidase I, α-fucosidase Os hydrolyzed both the α1,4-fucosyl linkage in Lewis a epitope of PCT N-glycans and the α1,3-fucosyl linkage in LNFP as shown in Fig. , but not the α1,3-fucosyl linkage in truncated type plant N-glycan (Xylβ1-2Manβ1-4GlcNAcβ1-4(Fucα1-3)GlcNAc-PA, MFXCitation9)) and Fucα 1-3GlcNAc-PA (derived from MFX by sequential enzymatic digestions). These results suggested that the Xylβ1-2Man unit or the β1,2-xylose residue might hamper the α1,3-fucosidase activity. At this moment, it is obscure whether α-fucosidase Os is preferentially active against the α1,4-fucosyl linkage or both the α1,4-fucosyl and the α1,3-fucosyl linkages in PTC N-glycans. Furthermore, we found that α-fucosidase Os was inactive against the pyridylaminated lacto-N-fucopentaose I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc-PA, Takara Bio Inc., Japan) (data not shown), indicating that α-fucosidase Os is one of typical α1,3/4 fucosidases. Sakurama et al. reported that two microbial α-fucosidases (Bacteriodes thetaiotaomicron enzyme (BT_2192)Citation10) and Bifidobacterium bifidum enzyme (BbAfcB)Citation11)) belong to GH29-B, which is active against α1,3/4-fucosyl linkage in Lewis x and Lewis a epitopes but not α 1,2-fucosyl linkage nor pNP-α-Fuc.Citation10) Based on the substrate specificity, rice α-fucosidase Os purified in this study seems to belong to GH29-B but not GH29-A containing α-fucosidases from Homo sapiense (FucA1 and A2), Drosophila melanogaster, and Lactobaccilus caseii (AlfA, AlfB, and AlfC),Citation10) although none of 3D structures of plant α-fucosidases being active against the complex type N-glycans has been determined. Furthermore, we found that α-fucosidase Os as well as the bacterial α-fucosidase (BT_2192) is inactive against Fucα1-3GlcNAc-PA, indicating that the protein structure of the substrate-binding site must be similar to each other. In the case of BT_2192, it has been found that a Gal-binding pocket consisted of W230, E254, and D277 plays a critical role for the hydrolytic activity against the two Lewis epitopes and the Fucα1-3(4)GlcNAc structure lacking of the Gal residue cannot be a substrate for BT_2192.Citation10) Therefore, we assume that α-fucosidase Os may have a similar sugar-binding pocket prerequisite for the α-fucosidase activity and the β1,2-Xyl-residue or the Xylβ1-2Man residues in MFX may hamper adequate accessibility of the α1,3 fucosyl residue to the catalytic site.
Based on the N-terminal amino acid sequence (A-A-P-T-P-P-P-L-) of the purified α-fucosidase Os, we performed a homology search using the BLAST program (NCBI, GenBank). This N-terminal sequence completely coincided with a part of the deduced amino acid sequence of one of putative rice α-fucosidases (Os04g0560400, putative α-fucosidase 1), which belongs to GH 29 family, as shown in Fig. . In the database, three putative GH29 family α-fucosidase genes (Os04g0560400, Os09g0520800, and Os10g0339290) were found in the phylogenic tree of the plant α-fucosidases (Fig. ), but two other α-fucosidases were not purified in this study. It is necessary, therefore, to purify and characterize these α-fucosidases, which may be encoded by Os09g0520800 and Os10g0339290, for understanding the defucosylation mechanism working in the turnover of plant complex N-glycans. Recently, we have identified a tomato (Solanum lycopersicum) α-fucosidase gene based on the N-terminal amino acid sequence of the present α-fucosidase Os and characterized the recombinant protein (α-fucosidase Sl, α-fucosidase from S. lycopersicum) expressed in the insect cells (Sf9).Citation12) Molecular cloning, detail analysis of substrate specificity, and the molecular modeling of α-fucosidase Sl will be described in our following paper.
Author contribution
Y.K. shared responsibility for the writing of the manuscript with M.Z.R, M.F., and M.M. All authors were responsible for the study concept and design. M.Z.R, M.F., and M.M carried it out. All authors contributed to the critical revision of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Program to Disseminate Tenure Tracking System from the Ministry of Education, Culture, Sports, Science and Technology; Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant number 24580494, 24580495].
Acknowledgments
This work was partially supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Basic Research (C), no. 24580494 to M.M. and no. 24580495 to Y.K.), and by grant from Program to Disseminate Tenure Tracking System (FY 2011-2013 to M.M.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Ogata-Arakawa M, Muramatsu T, Kobata A. α-L-fucosidases from almond emulsin: characterization of the two enzymes with different specificities. Arch. Biochem. Biophys. 1977;181:353–358.10.1016/0003-9861(77)90514-8
- Yoshima H, Takasaki S, Ito-Mega S, Kobata A. Purification of almond emulsin α-L-fucosidase I by affinity chromatography. Arch. Biochem. Biophys. 1979;194:394–398.10.1016/0003-9861(79)90632-5
- Imber MJ, Glasgow LR, Pizzo SV. Purification of an almond emulsin fucosidase on Cibacron blue-Sepharose and demonstration of its activity toward fucose-containing glycoproteins. J. Biol. Chem. 1982;257:8205–8210.
- Scudder P, Neville DC, Butters TD, et al. The isolation by ligand affinity chromatography of a novel form of α-L-fucosidase from almond. J. Biol. Chem. 1990;265:16472–16477.
- Zeleny R, Leonard R, Dorfner G, Dalik T, Kolarich D, Altmann F. Molecular cloning and characterization of a plant α1,3/4-fucosidase based on sequence tags from almond fucosidase I. Phytochemistry. 2006;67:641–648.10.1016/j.phytochem.2006.01.021
- Maeda M, Kamamoto M, Hino K, et al. Glycoform analysis of Japanese cedar pollen allergen, cry j 1. Biosci. Biotechnol., Biochem. 2005;69:1700–1705.10.1271/bbb.69.1700
- Maeda M, Kimura M, Kimura Y. Intracellular and extracellular free N-glycans produced by plant cells: occurrence of unusual plant complex-type free N-glycans in extracellular spaces. J. Biochem. 2010;148:681–692.10.1093/jb/mvq102
- Yokouchi D, Ono N, Nakamura K, Maeda M, Kimura Y. Purification and characterization of β-xylosidase that is active for plant complex type N-glycans from tomato (Solanum lycopersicum): removal of core α1-3 mannosyl residue is prerequisite for hydrolysis of β1-2 xylosyl residue. Glycoconjugate J. 2013;30:463–472.10.1007/s10719-012-9441-y
- Kimura Y, Minami Y, Tokuda T, Nakajima S, Takagi S, Funatsu G. Primary structures of N-linked oligosaccharides of momordin-a, ribosome inactivating protein from Momordica charantia seeds. Agric. Biol. Chem. 1991;55:2031–2036.10.1271/bbb1961.55.2031
- Sakurama H, Tsutsumi E, Ashida H, Katayama T, Yamamoto K, Kumagai H. Differences in the substrate specificities and active-site structures of two a-L-fucosidases (glycoside hydrolase Family 29) from Bacteroids thetaiotaomicron. Biosci. Biotechnol., Biochem. 2012;76:1022–1024.10.1271/bbb.111004
- Ashida H, Miyake A, Kiyohara M, et al. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017.10.1093/glycob/cwp082
- Rahman MdZ, Maeda M, Kimura Y. Molecular cloning and gene expression of α1,3,4-Fucosidase from Solanum lycopersicum. The 2015 Annual Conference of the Japan Society for Bioscience, Biotechnology and Agrochemistry; 2015 March 28; Okayama. Available from: http://www.jsbba.or.jp/2015; 3E32P17;78.
