Abstract
Disruption of two Fusarium genes that negatively regulate trichothecene biosynthesis was reported to cause a drastic increase in trichothecene production. However, careful inspection of these genes revealed that neither was significantly related to trichothecene production. Agmatine medium maintained the expression of trichothecene genes at significant levels, resulting in a 2–3-fold increase in the final yield, as compared to glutamine medium.
Trichothecenes are a large group of sesquiterpenoid mycotoxins produced by groups of fungi belonging to several distantly related genera such as Fusarium, Trichothecium, and Myrothecium.Citation 1 ) Among the fungal species that produce trichothecenes, Fusarium species impose the greatest impact on our economy and well-being because many are pathogens of important cereal crops such as wheat, maize, and barley.Citation 2 ) Fusarium graminearum and Fusarium culmorum are two such major pathogens; they are associated with the accumulation of type B trichothecenes such as deoxynivalenol (DON), nivalenol (NIV), and their O-acetyl derivatives that have a keto group at C-8. These Fusarium species produce trichothecenes under inducing conditions in liquid cultures. Although almost all the genes involved in the synthetic pathway were identified, the genetic elements involved in the regulatory network of trichothecenes remain to be clarified.Citation 3 )
Trichothecene production is regulated by a Cys2His2 zinc finger protein that is encoded by Tri6 Citation 4 ) residing at the central core region of the trichothecene gene cluster. The core region also contains the pathway gene Tri4 encoding a trichodiene monooxygenase responsible for the four consecutive oxygenation steps,Citation 1 ) and Tri5 encoding a trichodiene synthase responsible for the first cyclization.Citation 5 ) In addition to the trichothecene pathway and regulatory (Tri) genes, genes unrelated to trichothecene production were also found to be under the regulatory control of TRI6p.Citation 6 ) Among such gene products, FGSG_17598 (formerly registered as FGSG_00007; a putative cytochrome P450 enzyme) and FGSG_10397 (a terpene cyclase later identified as longiborneol synthase CLM1p), annotated by the Munich Information Center for Protein Sequences, are of particular interest because Gardiner and co-workersCitation 7 ) reported that deletion of either gene resulted in approximately a 14-fold increase in the amount of the mycotoxin produced, as compared to the wild-type (WT) strain. Increased metabolic flow to trichothecene biosynthesis adds value to the studies of trichothecene regulation as an attractive model system for efficient metabolic engineering of sesquiterpenes. However, the mechanism proposed for the overproduction of mycotoxin that results from gene disruption remains to be clarified. As the first step to understanding the regulatory system of trichothecenes, we re-examined the effect of disrupting the FGSG_17598 and FGSG_10397 genes on trichothecene production by using F. graminearum strain JCM 9873, a producer of trichothecene 15-acetyldeoxynivalenol (15-ADON).
Disruption mutants of FGSG_17598 and FGSG_10397 were generated by double-crossover homologous recombination. Vectors pJΔ17598 and pJΔ10397 were constructed for disruption of the FGSG_17598 and FGSG_10397 genes, respectively. These were created by placing a hygromycin B-resistant hph cassetteCitation 8 ) between the upstream and downstream regions of each gene by employing the inverse PCR (IPCR) method with the primers listed in Table S1. After linearization with NheI (see Fig. S1A), the disruption vectors were introduced into strain JCM 9873. Screening by PCR revealed that 4 and 9 out of 12 hygromycin B-resistant colonies for FGSG_17598 and FGSG_10397 genes, respectively, were the disruptants. Fig. S1B and Fig. S1C illustrate PCR and Southern blot analyses of representative strains of each deletion mutant, respectively. Theoretical sizes of the PCR amplicons and Southern bands, as expected from disruptive integration, were obtained for the two strains for each gene.
There were no marked differences in the morphology and color of the mycelia between the disruption mutant and WT strains. Production of trichothecenes by the FGSG_17598 and FGSG_10397 disruption mutants was examined using 30 mL of a glutamine or agmatine culture mediumCitation 9 ) in a 100 mL-Erlenmeyer flask shaken at 135 rpm (model NR-30 gyratory shaker; TAITEC Corp., Koshigaya, Japan) at 25 °C (see Table S2 for the exact composition of the media). Since a subsequent study revealed that agmatine and low pH synergistically enhance the expression of Tri5,Citation 10 ) the pH of the medium was not adjusted to pH 6.5.Citation 7 ) Similar to the 15-ADON/DON chemotype, strain JCM 9873 produced 15-ADON in liquid culture. The average 15-ADON level in glutamine medium (Table ) reached 59–71 μg/mL by day 8. Statistical analyses showed that the trichothecene level of the gene disruptants was similar to that of the WT strain. Trichothecene production by agmatine (120–178 μg/mL by day 8) was approximately 2–3-fold greater than by glutamine (Table ). Overall, disruption of the FGSG_17598 and FGSG_10397 genes did not result in a significant increase in the amount of 15-ADON.
Table 1. 15-ADON production by WT, FGSG_17598 disruption mutants Δ17598-#P2 and Δ17598-#P4, and FGSG_10397 disruption mutants Δ10397-#C1 and Δ10397-#C2 cultured in a synthetic liquid medium.
Because changes in the levels of the transcripts of the Tri genes and the genes for FGSG_17598 and FGSG_10397 were not monitored in the previous study,Citation 7 ) we examined the time course of their expression using fungal cultures with glutamine or agmatine. Expression analyses revealed circumstantial evidence suggesting the TRI6p-dependent expression of the FGSG_17598 and FGSG_10397 genes. The mRNA expression patterns of these genes were the same as those of Tri5 and Tri4 in the WT strain (Fig. ). The highest levels of expression were observed at 48 h, and then gradually decreased at 96 h in glutamine medium. Maximum mRNA levels of Tri5, Tri4, and FGSG_17598 and FGSG_10397 genes in agmatine medium at 48 h were generally equal to those in glutamine medium at the same time. However, medium composition exerted a major influence on the duration of gene transcription. Specifically, expression of these genes continued at 96 h in agmatine, but not in glutamine medium. Thus, the major medium contribution to trichothecene production was the long-lasting activation of the trichothecene gene, but not the maximum level of expression. Consistent with such an expression pattern, 15-ADON levels markedly increased after 48 h in agmatine medium as revealed by thin-layer chromatographic analysis (Fig. ). This could explain the greater accumulation of 15-ADON in agmatine than in glutamine medium.
Fig. 1. Northern blot analyses of FGSG_17598 and FGSG_10397 mRNA with reference to the onset of trichothecene accumulation in the medium as monitored by thin-layer chromatography (TLC).
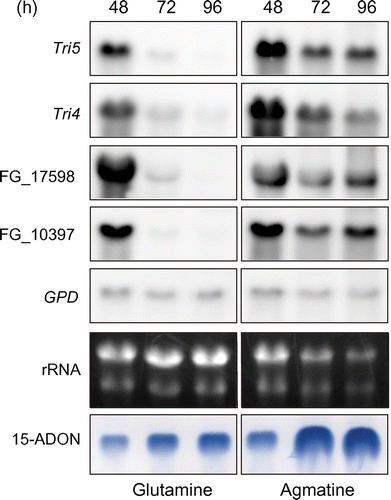
Neither of the genes studied were induced in the Tri6 disruption mutant ΔTri6 tk, which we generated in a previous study.Citation 11 ) No specific hybridization signals were detected on Northern blots (data not shown), and even sensitive reverse transcription (RT)-PCR analyses failed to detect these transcripts from the ΔTri6 tk mutant (Fig. S2). These results are consistent with a previous report on the features of FGSG_17598 and FGSG_10397 genes,Citation 7 ) suggesting conservation of the TRI6p-regulatory network of these genes in F. graminearum.
FGSG_17598 and FGSG_10397 show significant similarity to a cytochrome P450 monooxygenase and terpene cyclase, respectively.Citation 7 ) McCormick et al.Citation 12 ) demonstrated that the latter gene, designated CLM1 (FGSG_10397), encodes a longiborneol synthase. This enzyme is responsible for the first cyclization step in the biosynthesis of culmorin, another sesquiterpenoid secondary metabolite. Deletion of CLM1 from the genome of the F. graminearum strain 9F1 resulted in the complete disappearance of culmorin and seemingly increased production of 15-ADON, as revealed by GC-MS analysis.Citation 12 ) However, neither the amount of 15-ADON nor the level of Tri mRNA was analyzed in their study. A small increase in the amount of 15-ADON, if any, may be attributed to an increased availability of farnesyl pyrophosphate, a building block of sesquiterpenes for mycotoxin biosynthesis, because of the inactivation of a competing secondary metabolic pathway to culmorin.
In a previous report, 100 μL of synthetic liquid media distributed to 96-well plates were used for the fungal culture without shaking. By deleting either the FGSG_17598 or the FGSG_10397 gene, the amount of DON produced increased 14-fold with the agmatine medium.Citation 7 ) To exclude the possibility that a difference of culture conditions between a 100 mL-Erlenmeyer flask with shaking and a 96-well plate without shaking led to contradictory findings, we measured the amount of the mycotoxin that accumulated in 96-well plate cultures using strain JCM 9873. Although the concentration of 15-ADON was slightly increased in this system (maximum 206 μg/mL), marked differences in the amount of 15-ADON were not noticed with either medium between the WT and gene disruptants (Fig. S3).
In the previous study,Citation 7 ) F. graminearum CS3005, a strain that produces 15-ADON in liquid culture and DON in infected wheat heads, was used to assess the function of the FGSG_17598 and FGSG_10397 genes. The authors reported that DON production in both disruption mutants reached over 15 mg/mL in the agmatine medium (Table S2). This concentration is calculated to exceed 70% of the usable carbon source (agmatine plus sucrose) originally included in the agmatine medium. In wheat heads infected with the FGSG_17598 and FGSG_10397 disruption mutants, DON accumulates at a maximum level of 80 ppm and 250 ppm (mg toxin/kg plant material), respectively.Citation 7 ) Although the conversion rate of usable carbon sources to the product is difficult to calculate with live plants as a source of nutrients, the reported values of DON concentrations are again markedly increased by disruption of the FGSG_17598 and FGSG_10397 genes.Citation 7 )
With regard to the liquid culture, the previous studyCitation 7 ) used an ELISA kit (DON Plate kit ELISA; Beacon Analytical Systems Inc., Portland, ME, USA) to measure the toxin content. Since the antibody against DON shows 300% cross-reactivity to 15-ADON (manufacturer’s manual), the actual molecular species of type B trichothecene produced by the strain, and measured by the kit, is 15-ADON. Thus, the actual trichothecene concentration may be one-third of that described as DON in the previous publication.Citation 7 ) Even with this lower estimate, 5 mg/mL of 15-ADON indicates a yield of 23.3% based on sugar (carbon) consumption in the agmatine medium. In terms of carbon incorporation, this extremely high efficiency exceeds the yield of glutamic acid from glucose by Corynebacterium glutamicum no. 534 (ATCC 13032), an original strain isolated in 1956 in an attempt to develop a new glutamic acid production system in the fermentation industry.Citation 13,14 ) The negative regulatory system described in the previous studyCitation 7 ) may thus be highly specific to strain CS3005, and sesquiterpene regulation by a TRI6p-type transcription factor needs to be clarified by other clues using different approaches.
In summary, we re-examined the effects of disrupting FGSG_17598 (formerly registered as FGSG_00007) and FGSG_10397 in an attempt to understand the regulatory mechanisms of trichothecene production. As reported previously, their expressions were dependent on the function of the trichothecene cluster gene Tri6. However, in contrast to the previous report, neither of the deletion mutants produced significantly increased amounts of trichothecenes compared to the WT strain. While the mRNA level of Tri genes rapidly decreased in F. graminearum grown in glutamine medium, agmatine medium maintained the transcripts at significant levels even after a prolonged incubation period.
Authors contributions
Y.K., T.N., and M.K. designed the research plan; Y.K., Y.N., K.M., and Q.J. performed the experiments; Y.K., Y.N., and K.M. analyzed the data; K.K., T.K., and M.K. supervised the research; and M.K. wrote the manuscript.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2015.1088374.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry and JSPS KAKENHI [grant number 23658084].
Suppelementary_Figures.pdf
Download PDF (1.7 MB)Supplementary_Tables.docx
Download MS Word (47.2 KB)Additional information
Funding
References
- Kimura M , Tokai T , Takahashi-Ando N , et al . Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007;71:2105–2123.10.1271/bbb.70183
- Goswami RS , Kistler HC . Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004;5:515–525.10.1111/j.1364-3703.2004.00252.x
- Merhej J , Richard-Forget F , Barreau C . Regulation of trichothecene biosynthesis in Fusarium: recent advances and new insights. Appl. Microbiol. Biotechnol. 2011;91:519–528.10.1007/s00253-011-3397-x
- Proctor RH , Hohn TM , McCormick SP , et al . Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides . Appl. Environ. Microbiol. 1995;61:1923–1930.
- Hohn TM , Beremand PD . Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides . Gene. 1989;79:131–138.10.1016/0378-1119(89)90098-X
- Seong KY , Pasquali M , Zhou X , et al . Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009;72:354–367.10.1111/mmi.2009.72.issue-2
- Gardiner DM , Kazan K , Manners JM . Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol. Plant Microbe Interact. 2009;22:1588–1600.10.1094/MPMI-22-12-1588
- Staben C , Jensen B , Singer M , et al . Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newslett. 1989;36:79–81.
- Gardiner DM , Kazan K , Manners JM . Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 2009;46:604–613.10.1016/j.fgb.2009.04.004
- Gardiner DM , Osborne S , Kazan K , et al . Low pH regulates the production of deoxynivalenol by Fusarium graminearum . Microbiology. 2009;155:3149–3156.10.1099/mic.0.029546-0
- Nakajima Y , Tokai T , Maeda K , et al . A set of heterologous promoters useful for investigating gene functions in Fusarium graminearum . JSM Mycotoxins. 2014;64:147–152.10.2520/myco.64.147
- McCormick SP , Alexander NJ , Harris LJ . CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl. Environ. Microbiol. 2010;76:136–141.10.1128/AEM.02017-09
- Udaka S . Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus . J. Bacteriol. 1960;79:754–755.
- Udaka S . The dawning of amino acid fermentation (Review in Japanese). Kagaku To Seibutsu. 2009;47:211–216.10.1271/kagakutoseibutsu.47.211
- Takahashi-Ando N , Tokai T , Yoshida M , et al . An easy method to identify 8-keto-15-hydroxytrichothecenes by thin-layer chromatography. Mycotoxins. 2008;58:115–117.10.2520/myco.58.115
