Abstract
Analysis of expressed sequence tag libraries from various culture conditions revealed the existence of conidia-specific transcripts assembled to putative conidiation-specific reductase gene (csrA) in Aspergillus oryzae. However, the all transcripts were transcribed with opposite direction to the gene csrA. The sequence analysis of the transcript revealed that the RNA overlapped mRNA of csrA with 3′-end, and did not code protein longer than 60 amino acid residues. We designated the transcript Conidia Specific Long Natural-antisense RNA (CSLNR). The real-time PCR analysis demonstrated that the CSLNR is conidia-specific transcript, which cannot be transcribed in the absence of brlA, and the amount of CSLNR was much more than that of the transcript from csrA in conidia. Furthermore, the csrA deletion, also lacking coding region of CSLNR in A. oryzae reduced the number of conidia. Overexpression of CsrA demonstrated the inhibition of growth and conidiation, while CSLNR did not affect conidiation.
Graphical abstract
The results of EST analysis and Real-time PCR showed CSLNR is abundant in the conidia of Aspergillus oryzae. Deletion of both csrA and CSLNR occurred reduction of conidiation.
RNA is not always the intermediate product of protein. The vast majority of the human genome has been transcribed and it is known that less than 2% are genes that encode proteins.Citation1,2) RNAs not evident in protein coding are defined as noncoding RNAs. They can be divided into two groups: short non-coding RNAs and long non-coding RNAs.Citation3) Short non-coding RNAs include endogenous short interfering RNAs, like small interfering RNAs (siRNAs),Citation4) microRNAs (miRNAs),Citation4) and PIWI-interacting RNAs (piRNAs).Citation5,6) These short interfering RNAs form double-strand RNA, and induce RNAi.Citation3) Other classes of short non-coding RNAs include small nucleolar RNAs (SnoRNAs) involved in site-specific modification of transcripts,Citation7) and small nuclear RNAs (snRNAs) involved in transcription of target genes.Citation8,9) Long non-coding RNAs take part in diverse processes such as remodeling of chromatin,Citation8) altering protein activity,Citation9) and modulating protein location.Citation9)
In a subset of long non-coding RNA there are natural antisense RNA, of which transcriptional regions overlap with gene encoding proteins, and the RNA is transcribed with opposite direction to sense RNA.Citation10) Existence of natural antisense transcripts (NATs) in filamentous fungi has been reported by RNA-seq analysis in recent years. Of the 2,458 NATs found in Candida albicans, 10% overlapped with sense transcripts.Citation11) NATs are classified into three types, tail-to-tail type (3′ regions of sense RNA and antisense RNA are overlapped), head-to-head type (5′ regions of sense RNA and antisense RNA are overlapped), and embedded type (antisense RNA is embedded in sense RNA).Citation12) In Aspergillus flavus, RNA-seq identified 352 NATs: 46% tail-to-tail type, 37% embedded type.Citation12) In Aspergillus niger, NATs appear to be quite abundant in conidia and the regulation system of conidiation or germination by NATs was expected.Citation13) The function of these fungal NATs was uncertain and long non-coding RNAs were also unrevealed.
Conidia are important vehicles for deuteromycete, enabling a wide distribution of fungus. In species of Aspergillus, conidiation is also important development and conidia can be stored for more than a year.Citation14) In addition, conidia appear essential when Aspergillus is exposed to environmental stress. Conidiation was reported to be induced by nutrient starvation.Citation15) Moreover, proteins related with oxidative stress response were abundant in conidia.Citation16) Other reports indicated that proteins related with heat shock and dry stress responses were abundant in conidia.Citation17) In A. oryzae, several genes involved in conidiation were reported.Citation18,19) Disrupted mutants of the brlA gene formed immature conidiophore,Citation18) and the abaA deletion strain formed aberrant phialides.Citation19) The genes, fluG, flbA, flbB, flbC, flbD, flbE, and wetA are also reported to play roles in conidiation.Citation19) However, the whole picture of conidiation is not yet clear.
In A. oryzae, the existence of long non-coding RNA was not revealed previously. The results of expressed sequence tag (EST) analysis, sequence analysis and quantitative PCR revealed the existence of Conidia Specific Long Natural-antisense RNA (CSLNR) which overlapped with 3′- region of conidiation-specific reductase gene (csrA). We also demonstrated that deletion of csrA, which also lacked coding region of CSLNR and csrA resulted in reduction of conidiation. Overexpression of CsrA inhibited the growth and conidiation, while CSLNR did not affect conidiation.
Materials and methods
Strains and culture conditions
Strains used in this study are listed in Table . Wild type A. oryzae RIB40 was used for analyzing the transcriptional amounts of CSLNR and csrA in conidia, germinated conidia, and mycelia. Plasmid used for deletion of brlA was kindly gifted by Dr. Yamada (National Research Institute of Brewing).Citation18) A. oryzae NS4 strain derived from RIB40 was used for deletion of brlA as a host strain.Citation20) In the construction of the csrA deletion strain, A. oryzae ΔligD::ptrA was used to improve gene targeting.Citation21) Escherichia coli DH5α cells and Saccharomyces cerevisiae BY4741 cells were used for gene manipulation.
Table 1. Strains used in this study.
Czapeck-Dox (CDN) medium (3% glucose, 0.6% NaNO3, 0.052% KCl, 0.0815% KH2PO4, 0.1045% K2HPO4, 0.052% MgSO4, 0.1% trace element solution (0.1% FeSO4·7H2O, 0.88% ZnSO4·7H2O, 0.04% CuSO4·5H2O, 0.01% Na2B4O7·10H2O, 0.005% (NH4)6Mo7O24·4H2O), 2% agar) was used for culturing RIB40, ∆csrA-pNGA142, ΔRIB40, ΔcsrA-pNGA142 csrA and CSLNR overexpression strain, selection of csrA and CSLNR over expression strain. For culturing csrA and CSLNR over expression strain under induction condition, 2% maltose and 0.15% L-methionine and 1.03% L-glutamate were added to CDN medium instead of glucose and NaNO3 (CDML). Instead of maltose, fructose was added to CDML medium (CDFL) for culturing csrA and CSLNR overexpression strain under non-induction condition. NaNO3 was removed and 0.15% L-methionine and 1.03% L-glutamate were added to CDN medium (CDNL) for culturing ΔligD::ptrA and ΔcsrA strains. CD medium, which removed NaNO3 and added L-glutamate (CDL) was used for selection of ΔcsrA and ΔbrlA strains. NS4 strain and ΔbrlA strain were cultured on potato dextrose agar (PD) medium to which 1% agar was added (3.9% Potato Dextrose Agar (Nippon Becton, Dickinson and Company, Tokyo, Japan), 1% agar). All inoculated media were incubated at 30 °C.
Construction of the csrA deletion strain
The csrA deletion strain was constructed as follows, according to the standard method described by Ausubel.Citation22) The deletion cassette for csrA was cloned into pYES2 yeast expression vector (Thermo Fisher Scientific K.K., Yokohama, Japan). The upstream region of the csrA gene (1.3 kb) was amplified by PCR with a csrAupY2F primer and a csrAupY2R primer employing ΔligD::ptrA genome as a template. The downstream region of the csrA gene (1.4 kb) was amplified by a csrAdownY2F primer and a csrAdownY2R primer, employing ΔligD::ptrA genome as a template. The gene, sC in pUSCCitation20) was amplified with a pair primers of sCY2F and sCY2R. Then, pYES2 was digested with BamHI and EcoRI. Fragments of csrA upstream region, csrA downstream region, sC ORF, and digested pYES2 were introduced into S. cerevisiae BY4741 to ligate the fragments by the lithium acetate method.Citation23) The resulting plasmid was named pcsrA-sC. The deletion cassette for csrA was amplified by PCR employing csrAupY2F and csrAdownY2R as primers and pcsrA-sC as a template. Transformation was performed as described in a previous studyCitation24) and the cassette for csrA deletion was introduced into the A. oryzae ΔligD::ptrA strain. The deletion of csrA gene was confirmed by Southern blotting using two DNA fragments as probes. Probe A was generated by PCR using sCprobeF and sCprobeR as primers and genome DNA of A. oryzae RIB40 as a template. Probe B was generated in the same manner using csrAupstreamF and csrAupstreamR (Table ).
Table 2. Primers used in this study.
Construction of csrA overexpression strain
A plasmid pNGA142 was used as an expression vector for constructing csrA overexpression strain, CSLNR overexpression strain.Citation25) First, the ORF region of csrA was amplified by PCR, employing csrAOEF primer and csrAOER primer as primers and A. oryzae niaD300Citation26) genome DNA as a template. The transcriptional region of CSLNR was amplified by PCR, employing cslnrOEF and cslnrOER as primers and A. oryzae niaD300 genome DNA as a template. The PCR fragments were digested with SpeI and HindIII and inserted into SpeI-HindIII digested pNGA142. The resulting plasmids and pNGA142, digested with MunI, was introduced into ∆csrA. The construction of csrA and CSLNR overexpression starin was confirmed by Southern blotting using DNA fragments as probe C. Probe C was generated by PCR using niaDprobeF and niaDprobeR as primers and genome DNA of A. oryzae niaD300 as a template.
Harvest of conidia, germinated conidia, mycelia
We used A. oryzae RIB40 strain for transcriptional analysis. RIB40 strain was inoculated on CDN agar medium and cultivated at 30 °C. Plates were incubated for 8 days. Conidia were separated with a 70-μm pore-size cell strainer (Nippon Becton, Dickinson and Company, Tokyo, Japan), harvested with 10 mL of sterilized distilled water (SDW), and then 5 μL of conidia suspension (8.8 × 106 conidia/mL) was inoculated on CDN agar medium again and cultured at 30 °C for 11 days. Conidia were harvested with 10 mL of 0.01% polyoxyethylene (20) sorbitan monolaurate solution, separated by a 70-μm pore-size cell strainer, and then washed twice with SDW. For harvesting germinated conidia, 3.0 × 108 conidia prepared as described above were suspended into 100 mL of CDN liquid medium and cultivated for 2, 6, and 8 h at 30 °C. Cultivated medium was transferred into a conical centrifuge tube (Nippon Becton, Dickinson and Company, Tokyo, Japan). Polyoxyethylene (20) sorbitan monolaurate was added to the culture broth, achieving a final concentration of 0.02%. The solution was centrifuged at 8340 × g for 20 min and the germinated conidia were collected. For harvesting mycelia, 1.0 × 108 conidia prepared as described above were inoculated into 100 mL of CDN liquid medium and cultivated at 30 °C for 24 and 48 h. Mycelia were harvested with Miracloth (Merk Japan Ltd., Tokyo, Japan). Harvested conidia, germinated conidia, and mycelia were frozen with liquid nitrogen and stored at −80 °C.
Total RNA preparation, cDNA synthesis, real-time PCR
For RNA preparation, all samples were frozen with liquid nitrogen and ground with mortar and pestle to powder. All molecular manipulations were performed according to the manufacturers’ instructions. Total RNA was isolated using Sepazol-RNA I (Nacalai Tesque Inc., Kyoto, Japan). DNase treatment was conducted using RNase-free Recombinant DNase I (Takara Bio Inc., Kusatsu, Japan) to eliminate genome DNA. To obtain cDNA pools from total RNA, a reverse transcriptase reaction was performed using PrimeScript™ II 1st strand cDNA Synthesis Kit (Takara Bio Inc, Kusatsu, Japan). Two hundred nanograms of total RNA was used as a template, and random 6-mer primers from the kit were used as reverse transcriptase primer. Real time PCR was conducted using SYBR Premix Ex Taq™II (Takara Bio Inc, Kusatsu, Japan) with Mx3005P QPCR (Agilent technologies Inc, Santa Clara, CA, USA) using conditions consisting of 30 s at 95 °C followed by 40 cycles of 5 s at 95 °C, 30 s at 64 °C. Primers, CSLNRrealtimeF2 and CSLNRrealtimeR2 were used to detect CSLNR. Primers, csrArealtimeF and csrArealtimeR were used to detect transcript from csrA. Each primer set was designed to amplify 5′ ends of CSLNR and the transcript from csrA to detect each transcript, respectively. Transcriptional regions of the csrA were determined from the A. oryzae RNA seq database CAoGD (http://nribf21.nrib.go.jp/CFGD/). The 5′ and 3′ ends of CSLNR were confirmed by 5′- and 3′-RACE, respectively. The primer set of actinrealtimeF and actinrealtimeR was used to amplify the β actin gene.
Sequence analysis of CSLNR and csrA
Sequence analysis of CSLNR was conducted by employing total RNA from RIB40. Complementary DNA pools were obtained from total RNA. A reverse transcriptase reaction was performed using a PrimeScript™ II 1st strand cDNA Synthesis Kit with oligo dT primer (Takara Bio Inc, Kusatsu, Japan). Transcriptional region of CSLNR was amplified by PCR, employing an F1ecoRI primer and an R1speI primer. The PCR fragments were digested with EcoRI and SpeI, and inserted into EcoRI-SpeI digested pBluescripII KS(+). The nucleotide sequence of the inserted fragment was confirmed using an ABI 310 Genetic Analyzer (Applied Biosystems Inc, Foster City, CA, USA), employing pblue550F and pblue800R primers.
Five prime and three prime ends of CSLNR were confirmed by 5′- and 3′-rapid amplification of cDNA ends (RACE) methods, respectively. The 5′-RACE methods were conducted using 5′-Full RACE core set (Takara Bio Inc, Kusatsu, Japan), and 5dRT2 was used as the reverse transcriptase primer. The first PCR was performed employing 5dF6 and 5dR6 as the primers, and a second PCR was performed employing primers, 5dF7 and 5dR7. Subcloning PCR fragments into pBluescriptII KS(+) and sequencing were performed as described above. To perform 3′-RACE, TaKaRa RNA PCR kit (AMV) Ver. 3.0 was used. Oligo dT-adapter primer was used as a reverse transcriptase primer. The primers, M13M4 and 5dantR2 primer were employed for PCR. The PCR fragment was subcloned into pGEM-T easy vector (Promega KK, Tokyo, Japan). Sequencing was performed as described above.
Sequence analysis of csrA was conducted by employing total RNA from csrA over expression strain. Complementary DNA pool was obtained as described above. In order to obtain transcript from csrA, PCR was conducted employing the primer set of csrAOEF and csrAOER. The PCR fragments were digested with HindIII and SpeI, and inserted into HindIII-SpeI digested pBluescriptII KS (+).
Morphology observation
Conidial suspensions of each strain (2.0 × 105/mL) was inoculated on CDNL, CDFL, CDML agar medium, and grown at 30 °C for 5 days. Fungal growth was evaluated as colony diameter. The numbers of spores were counted by hemacytometer after preparing conidial suspension as described above.
Results
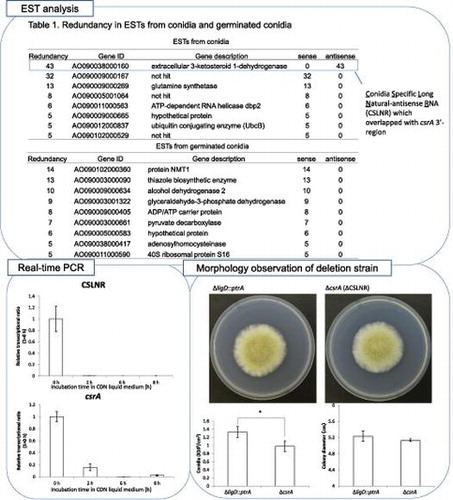
EST analysis was conducted to analyze the transcriptional landscape of A. oryzae in various culture conditions as reported in a previous study.Citation27) At first, we conducted sequence assembly of the raw ESTs from liquid germinated conidia and conidia (LG) library. We revealed that some of these tags showed redundancy. Eight genes assembled more than five ESTs from conidia, and nine genes assembled more than five ESTs from germinated conidia (Table ). We quoted the gene description from NCBI (http://www.ncbi.nlm.nih.gov). In particular, 43 tags from conidia were assembled to AO090038000160 (Table , Fig. ). The transcripts were identified only in conidia, and any tags assembled to AO090038000160 were not found in germinated conidia. In addition, all of assembled tags were derived from transcripts that were transcribed with opposite direction to predicted ORF, AO090038000160 by DOGAN (http://www.bio.nite.go.jp/dogan/project/view/AO), which we named conidiation-specific reductase (csrA). The sequence of complementary DNA (cDNA) from csrA was shown in Fig. . The result indicated that the putative exons and introns of AO090038000160 were differnt from those of actual csrA. We revealed that the csrA gene had five introns and could code a protein consisting of 444 amino acid residues (accession number; LC033559). The deduced protein contained a catalytic domain conserved in short-chain dehydrogenases/reductases (SDRs) and an NAD binding domain.
Table 3. Redundancy in ESTs from conidia and germinated conidia ESTs from conidia.
Fig. 1. Expressed sequence tags (ESTs) from conidia assembled to csrA. Right arrow indicates transcriptional direction of csrA, left arrow indicates transcriptional direction of CSLNR.
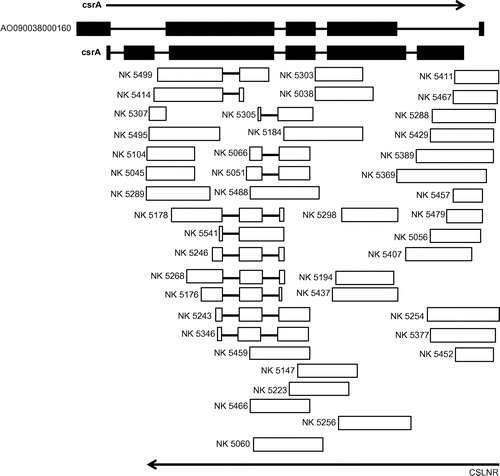
Fig. 2. Nucleotide sequence of csrA gene region of Aspergillus oryzae.
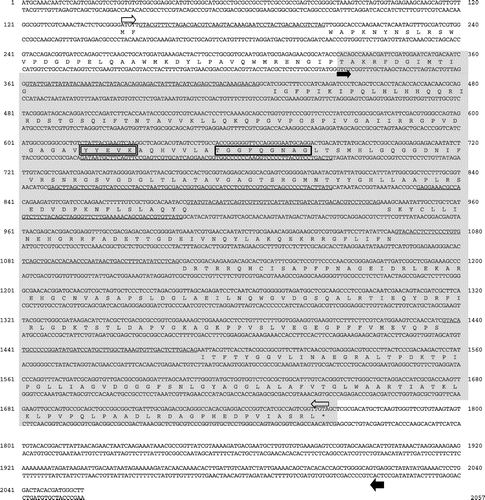
Next, we performed 5′- and 3′-RACE of the transcripts which were transcribed in the opposite direction to the gene csrA. We revealed that 5′ end of the transcripts were located 253 bp downstream of the open reading frame of the csrA gene. Two 3′ ends of the transcripts were indicated at 194 and 202 bp of csrA gene. The transcripts were found to be spliced at three locations (Fig. ). Three splicing patterns were observed: only the third intron from 5′ end of the transcripts was spliced; the first intron and the third introns were spliced; all introns were spliced. The length of the transcripts whose introns were all spliced was 1,492 base. Analyzing the transcriptional end site of the transcripts by 3′ RACE method, we employed Oligo dT-adapter primer as a reverse transcriptase primer, indicating that the 3′-end of the transcripts were polyadenylated. In all splicing patterns and codon frames, the transcripts could not code peptides longer than 60 amino acid residues. We designated the transcripts as CSLNR. The results indicated that the transcriptional region of the gene csrA and CSLNR overlapped with their 3′-ends.
Then we analyzed the transcriptional amount of CSLNR and csrA. To distinguish transcripts from CSLNR and csrA, we designed the qPCR primers (csrArealtimeF primer, csrArealtimeR primer, cslnrrealtimeF primer, cslnrrealtimeR primer) which amplified 5′ ends of CSLNR and csrA mRNA specifically. Transcriptional regions of csrA were quoted from CAoGD (http://nribf21.nrib.go.jp/CFGD/). We compared the transcriptional amount of CSLNR and csrA in conidia (0 h) and mycelia (24 and 48 h after incubation). Both CSLNR and csrA mRNA were observed only in conidia, and they were nearly absent in mycelia (Fig. ). Then, we analyzed the change of transcriptional amount of CSLNR and csrA during germination. Swollen conidia were observed after 2 h, and germination started at 6 h (Fig. (A)). Germinated conidia were observed after incubation for 8 h (Fig. (A)). Results of real-time PCR indicated that CSLNR was abundant only in non-incubated conidia, and was not detected in conidia incubated more than 2 h. Meanwhile, the results also indicated that transcripts from csrA were abundant in conidia, and decreased to 16% of RNA after incubation for 2 h. The transcript from csrA was not observed in germinated conidia (Fig. (B)). Comparing the abundance of transcripts from CSLNR and csrA in conidia (0 h), we observed that the transcriptional level of CSLNR was more than 4,500 times higher than that of csrA in conidia (Fig. (C)).
Fig. 3. Transcriptional analysis of CSLNR and csrA in conidia and mycelia.
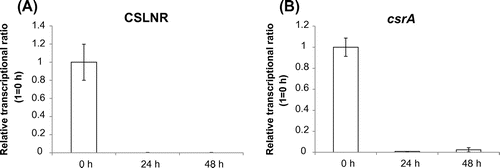
Fig. 4. Transcriptional analysis of CSLNR and csrA in conidia and germinated conidia.
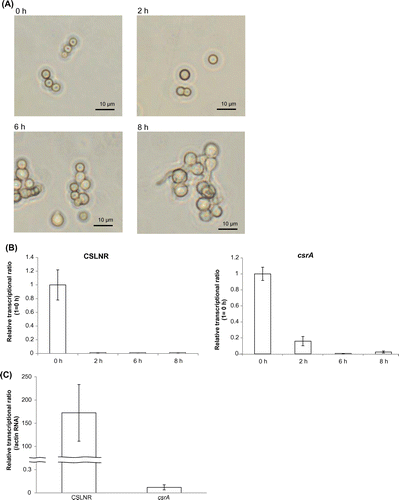
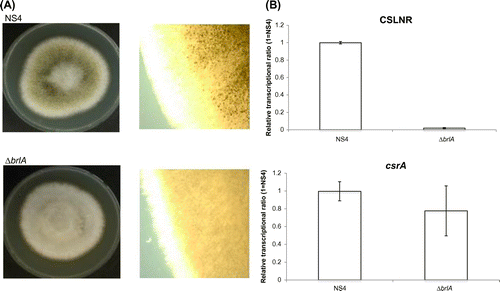
We analyzed the amount of both RNA in A. oryzae brlA deletion strain, which was unable to form mature conidiophores, by misconducting the development of conidiophore structuresCitation18,28) to reveal when the transcription of CSLNR and the gene csrA started. The host strain, A. oryzae NS4, which was derived from RIB40, was used as the control in this study. brlA deletion strain and NS4 strain were cultured on PD medium at 30 °C for 8 days. Conidiation was observed in NS4 but not observed in brlA deletion strain as previously reported (Fig. (A)).Citation18) Next, we conducted transcriptional analysis of CSLNR and csrA in NS4 and ΔbrlA strain. NS4 and ΔbrlA strains were inoculated on PD medium. After incubating at 30 °C for 7 days, total RNA was extracted from each strain and used in RT-PCR. Results of real-time PCR indicated that the abundance of the transcript from csrA did not change by deletion of brlA, but the abundance of CSLNR drastically reduced to 1.9% of NS4 (Fig. (B)). These results suggested that CSLNR could not be transcribed in the absence of brlA, and transcriptional patterns differed between CSLNR and csrA. The results indicated that the transcription of CSLNR was conidia specific and depends on brlA, and then, vanished at swelling of conidia completely.
Fig. 5. Transcriptional analysis of CSLNR and csrA in NS4 strain and ΔbrlA.
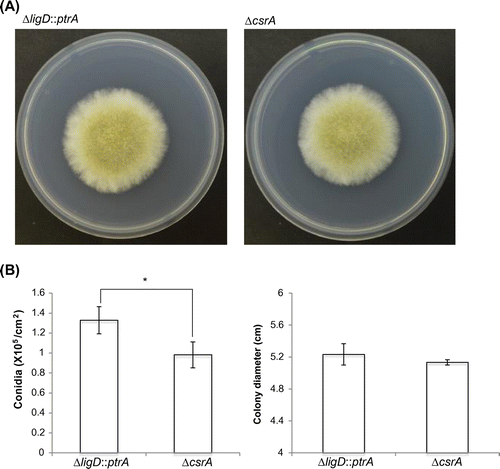
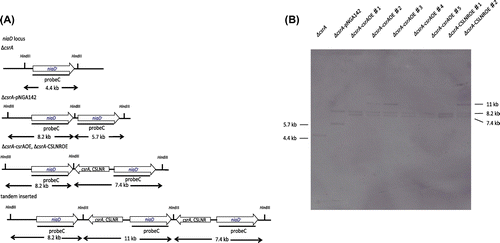
We constructed the deletion strain of csrA. It also lacks the coding sequence of CSLNR 3′- overlapping region. The results of Southern blot hybridization are displayed in Fig. (A) and (B). The SalI fragments of 6.7 and 2.8 kb were detected by probe A in ΔligD::ptrA and ΔcsrA. The PstI fragments of 7.8 and 9.2 kb were detected by probe B in ΔligD::ptrA and ΔcsrA, respectively. These results indicated that deletion cassette of csrA was inserted into csrA locus only. In comparision between ∆csrA strain and control strain, colony diameter did not change. However, the ability of conidiation was slightly decreased in ΔcsrA (Fig. (A) and (B)).
Fig. 6. Confirmation of ΔcsrA by Southern blot hybridization analysis.
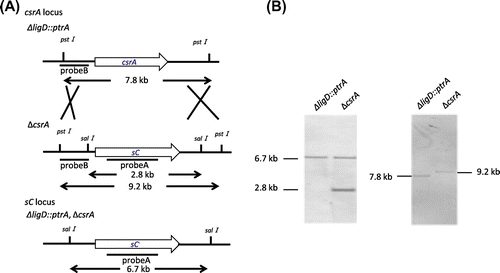
Fig. 7. Phenotypic analysis of ΔcsrA.
Next, we constructed csrA and CSLNR overexpression strain employing ∆csrA as a host strain to check which of CsrA and CSLNR promote conidiation. We obtained five candidates of csrA overexpression strain (∆csrA-csrAOE♯1-5) and two candidates of CSLNR overexpression strain (∆csrA-CSLNROE♯1-2). The results of Southern blot hybridization, which using probe C, are displayed in Fig. (B). The HindIII fragments of 4.4 kb were detected by probe C in niaD300. The HindIII fragments of 5.7, 8.2 kb were detected by probe C in niaD300-pNGA142. The HindIII fragments of 7.4, 8.2 kb were detected by probe C in ∆csrA-csrAOE♯1, ΔcsrA-csrAOE♯4 and ΔcsrA-csrAOE♯5, and ΔcsrA-CSLNROE♯1. These results indicated that the fragments for construction of csrAOE and CSLNROE strains were inserted singly in ∆csrA-csrAOE♯1, ΔcsrA-csrAOE♯4 and ΔcsrA-csrAOE♯5, and ΔcsrA-CSLNROE♯1, respectively. The HindIII fragments of 7.4, 8.2 and 11 kb were detected by probe C in ΔcsrA-csrAOE♯2 and ΔcsrA-csrAOE♯3, and ΔcsrA-CSLNROE♯2, respectively. These results indicated that the fragments for construction of csrAOE and CSLNROE strain were inserted tandemly in ΔcsrA-csrAOE♯2 and ΔcsrA-csrAOE♯3, and ΔcsrA-CSLNROE♯2, respectively. We employed ΔcsrA-csrAOE♯5 andΔcsrA-CSLNROE♯1 for the further experiments.
Fig. 8 Confirmation of ∆csrA-csrAOE, ∆csrA-CSLNROE by Southern blot hybridization analysis.
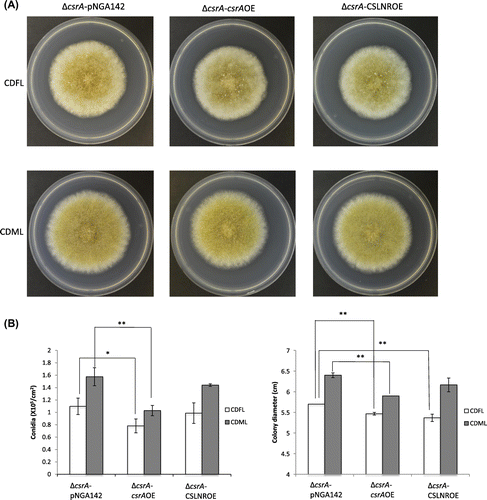
The overexpression strains were grown on CDFL medium and CDML medium at 30 °C for 5 days. It has been reported that glaA promoter is induced in the presence of maltose.Citation29) Both of the overexpression of CSLNR and csrA did not promote conidiation, though the deletion both of CSLNR and csrA showed conidiation decrease. Meanwhile, the overexpression of csrA inhibited the conidiation and growth in both CDFL and CDML medium. On the other hand, the overexpression of CSLNR did not affect conidiation, but inhibited the growth on both CDFL and CDML medium (Fig. (B)).
Fig. 9. Phenotypic analysis of ∆csrA-pNGA, ∆csrA-csrAOE and ∆csrA-CSLNROE.
Discussion
The results of EST analysis demonstrated that some genes were expressed with high frequency in conidia, but it was unclear how these genes were linked to conidiation because their functions were unproven.Citation27) Among the genes, CSLNR showed the highest redundancy in conidia. Unlike other abundant transcripts in conidia, CSLNR were transcribed with the opposite direction of csrA predicted by genomic analysis.Citation30) As mentioned in the results, sequence analysis and 5′- and 3′-RACE of CSLNR indicated that CSLNR could not code any protein in spite of csrA coding the protein of 444 amino acid residues. In the previous report, it was suggested that the average length of 5′ and 3′ UTR in fungi are 134.0 and 237.1 bp, respectively.Citation31) If the peptide of 60 amino acid residues of peptide was translated from CSLNR, CSLNR is too long (5′ UTR: 577 bp, 3′ UTR: 737 bp). We searched homologous peptides and domains against the putative 60 amino acid residues peptide using blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Pfam database (http://pfam.xfam.org/), but no homologous peptide and domain were found. Based on these results, CSLNR existing dominantly in conidia appeared not to be translated into peptide and to work as the RNA.
The results of the real-time PCR indicated that the transcription of CSLNR was vanished in the absence of brlA, although the abundance of transcripts from csrA did not change in ∆brlA strain compared to that in NS4 strain. In the previous reports, BrlA response element (BRE) has been identified.Citation32) We searched the sequences of BRE in 2,000 bp upstream of csrA ORF and 5′ end of CSLNR. BRE was found at 190 bp upstream from the start codon of csrA, but not found in 2,000 bp upstream region of CSLNR. By these results, the expression of csrA and CSLNR would be independent of BrlA. Next, we searched the sequence of the AbaA binding siteCitation33) in the same upstream region of csrA and CSLNR. The sequence of AbaA binding sites were found at 810 and 824 bp upstream of the start codon of csrA and 913 and 546 bp upstream of the 5′ end of CSLNR. The abundance of CSLNR was absolutely rare in ∆brlA strain. The transcription of abaA depends on brlA,Citation19) so the transcription of CSLNR might be activated by AbaA. Otherwise, the transcription of csrA and CSLNR might be regulated by fluG, flbB, flbC, flbD, flbE, which were reported to be related with conidiation.Citation19)
The defects of CsrA and CSLNR reduced conidiation. However, It was unclear which defect of CsrA or CSLNR was concerned with conidiation reduction. We constructed csrA overexpression strain, ∆csrA-csrAOE and CSLNR overexpression strain, ∆csrA-CSLNROE using ∆csrA strain as hosts to check which of CsrA and CSLNR was able to promote conidiation. We thought the reduction of conidiation in ∆csrA would be owing to the defect of CSLNR, because the transcriptional level of csrA was much lower than that of CSLNR in conidia. In conidia, the transcript from csrA was most abundant, the transcriptional level of csrA was 14.2 times lower than that of β-actin gene (data not shown). On the other hand, the transcriptional level of CSLNR was 172.4 times higher than that of β-actin gene in conidia. Because the transcriptional level of csrA was very low as described above, we think the impact of the defect of the gene csrA was very small. However, the overexpression of CSLNR could not promote conidiation (Fig. (B)). As the result of it, it seems that CSLNR is unable to promote conidiation individually.
The overexpression of csrA inhibited the growth and conidiation in CDFL and CDML medium. The transcription of csrA was not observed in wild type mycelia naturally, but we employed glaA promoter that was strongly induced in myceliaCitation34) for the expression of CsrA. The growth of mycelia might be worse for unnecessary CsrA function. Furthermore, the transcriptional level by glaA promoter would be higher than that of csrA promoter. It would bring out during conidia formation. The transcriptional level of csrA was very low as described above even in conidia (Fig. ). The excessive CsrA in conidia might disorder the conidia formation. On the other hand, the absence of CSLNR might cause the inhibition of conidiation. In Aspergillus genus, it was reported that the presence of antisense RNA is able to repress expression of gene. In the previous report, overexpressing antisense RNA of pepB reduced the PepB protein level ranged between 10 and 70% in the every transformant of A. awamori.Citation35) The results of real-time PCR demonstrated that csrA mRNA existed in conidiophore without conidia, conidia and swelled conidia. Meanwhile, CSLNR was conidia specific (Fig. ). Considering the facts, the abundant transcription of CSLNR might be necessary to repress the expression of CsrA in the late stage of conidiation.
With all these factors, CsrA seems to be concerned with the conidiation of A. oryzae with adequate quantity and in a fine timing, and CSLNR might contribute to orchestrate the quantity of CsrA during conidiation.
We found that the homologous genes of csrA are present in A. niger and A. flavus; An08g04730 and AFL2G_08972, respectively. The presence of antisense RNA of An08g04730 and AFL2G_08972 has not been reported. Novodvorska et al. described the presence of NATs was detected in the conidia of A. niger.Citation13) However, the transcript from An08g04370 was not detected in conidia, differently from CSLNR and csrA, which were abundant in the conidia of A. oryzae. Meanwhile antisense transcripts have been reported in the mycelia and sclerotia in A. flavus,Citation36) but the antisense transcript of AFL2G_08972 was not reported in the previous paper.
The significance of long natural antisense RNA in Aspergillus genus remains undescribed. CSLNR has not been found in the other Aspergillus, and this is the first paper that reports the accumulation of CSLNR in the conidia of A. oryzae.
Authors contributions
Y. Yamagata, M. Takeuchi and M. Tsujii designed the study. K. Ishi, K. Madokoro, S. Okuda, M. Tsujii performed research. Y. Yamagata and M. Tsujii wrote the manuscript. All authors reviewed and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgments
We thank Dr. Yamada (National Research Institute of Brewing) for plasmid used for deletion of brlA.
References
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–299.10.1002/(ISSN)1521-1878
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–81610.1038/nature05874
- Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139.10.1002/path.v220:2
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655.10.1016/j.cell.2009.01.035
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103.10.1016/j.cell.2007.01.043
- Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011;90:430–440.10.1093/cvr/cvr097
- Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790.10.1016/S0300-9084(02)01402-5
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227.10.1038/nature07672
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504.10.1101/gad.1800909
- Osato N, Suzuki Y, Ikeo K, et al. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306.
- Tuch BB, Mitrovich QM, Homann OR, et al. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070.10.1371/journal.pgen.1001070
- Smith CA, Robertson D, Yates B, et al. The effect of temperature on Natural Antisense Transcript (NAT) expression in Aspergillus flavus. Curr. Genet. 2008;54:241–269.10.1007/s00294-008-0215-9
- Novodvorska M, Hayer K, Pullan ST, et al. Transcriptional landscape of Aspergillus niger at breaking of conidial dormancy revealed by RNA-sequencing. BMC Genomics. 2013;14:246.10.1186/1471-2164-14-246
- Lamarre C, Sokol S, Debeaupuis JP, et al. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics. 2008;9:417.10.1186/1471-2164-9-417
- Skromne I, Sánchez O, Aguirre J. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology. 1995;141:21–28.
- Suh MJ, Fedorova ND, Cagas SE, et al. Development stage-specific proteomic profiling uncovers small, lineage specific proteins most abundant in the Aspergillus fumigatus conidial proteome. Proteome Sci. 2012;10:3010.1186/1477-5956-10-30.
- van Leeuwen MR, Krijgsheld P, Bleichrodt R, et al. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud. Mycol. 2013;74:59–70.10.3114/sim0009
- Yamada O, Lee BR, Gomi K, et al. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J. Biosci. Bioeng. 1999;87:424–429.10.1016/S1389-1723(99)80089-9
- Ogawa M, Tokuoka M, Jin FJ, et al. Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 2010;47:10–18.10.1016/j.fgb.2009.10.004
- Yamada O, Lee BR, Gomi K. Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosei. Biotech. Biochem. 1997;61:1367–1369.10.1271/bbb.61.1367
- Mizutani O, Kudo Y, Saito A, et al. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 2008;45:878–889.10.1016/j.fgb.2007.12.010
- Ausubel, MF, Brent R, Kingston RE, et al. Current protocols in molecular biology. New York: 1987.
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96.10.1016/S0076-6879(02)50957-5
- Morita H, Abo H, Okamoto A, et al. Enzymatic properties of the recombinant serine-type carboxypeptidase OcpC, which is unique to Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2011;75:662–668.10.1271/bbb.100749
- Minetoki T, Tsuboi H, Koda A, et al. Development of high expression system with the improved promotor using the cis-acting element in Aspergillus species. J. Sci. Soc. Biol. Macromol. 2003;3:89–96.
- Minetoki T, Nunokawa Y, Gomi K, et al. Deletion analysis of promoter elements of the Aspergillus oryzae agdA gene encoding alpha-glucosidase. Curr. Genet. 1996;30:432–438.10.1007/s002940050153
- Akao T, Sano M, Yamada O, et al. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 2007;14:47–57.10.1093/dnares/dsm008
- Yamada O, Ikeda R, Ohkita Y, et al. Gene silencing by RNA interference in the koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007;71:138–144.10.1271/bbb.60405
- Hata Y, Kitamoto K, Gomi K, et al. Functional elements of the promoter region of the Aspergillus oryzae glaA gene encoding glucoamylase. Curr. Genet. 1992;22:85–91.10.1007/BF00351466
- Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161.10.1038/nature04300
- Pesole G, Mignone F, Gissi C, et al. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81.10.1016/S0378-1119(01)00674-6
- Chang YC, Timberlake WE. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1993;133:29–38.
- Andrianopoulos A, Timberlake WE. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell. Biol. 1994;14:2503–2515.10.1128/MCB.14.4.2503
- Minetoki T, Kumagai C, Gomi K, et al. Improvement of promoter activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl. Microbiol. Biotechnol. 1998;50:459–467.10.1007/s002530051321
- Moralejo FJ, Cardoza RE, Gutierrez S, et al. Silencing of the aspergillopepsin B (pepB) gene of Aspergillus awamori by antisense RNA expression or protease removal by gene disruption results in a large increase in thaumatin production. Appl. Environ Microbiol. 2002;68:3550–3559.10.1128/AEM.68.7.3550-3559.2002
- Wu X, Zhou B, Yin C, et al. Characterization of natural antisense transcript, sclerotia development and secondary metabolism by strand-specific RNA sequencing of Aspergillus flavus. PLoS One. 2014;9:e97814.10.1371/journal.pone.0097814
