Abstract
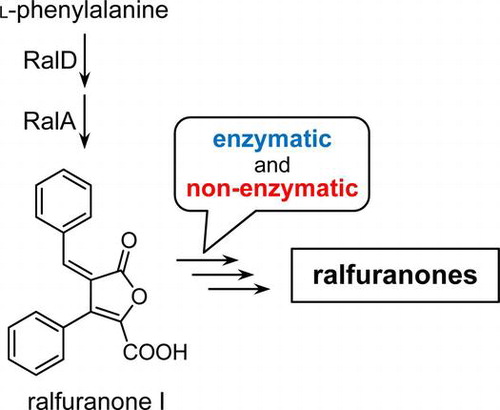
Ralfuranones, aryl-furanone secondary metabolites, are involved in the virulence of Ralstonia solanacearum in solanaceous plants. Ralfuranone I (6) has been suggested as a biosynthetic precursor for other ralfuranones; however, this conversion has not been confirmed. We herein investigate the biosynthesis of ralfuranones using feeding experiments with ralfuranone I (6) and its putative metabolite, ralfuranone B (2). The results obtained demonstrated that the biosynthesis of ralfuranones proceeded in enzymatic and non-enzymatic manners.
Graphical abstract
Proposed biosynthetic pathway of ralfuranones in Ralstonia solanacearum.
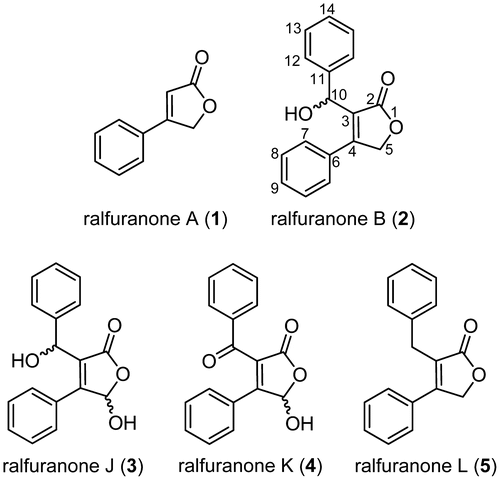
Ralstonia solanacearum is a soil Gram-negative bacterium that causes “bacterial wilt” on a wide range of host plants, including economically important crops such as the tomato, tobacco, potato, peanut, and banana.Citation1) The bacterium invades the intercellular spaces of roots through openings such as wounds, accumulates around the stele, and then breaks into and fills xylem vessels through the action of cellulolytic enzymes on vessel walls.Citation2) The ability of this pathogen to cause host wilting has mainly been attributed to its production of extracellular polysaccharide (EPS).Citation3) The production of EPS was previously shown to be regulated by quorum sensing (QS) consisting of phc regulatory elements, in which the global virulence regulator PhcA plays a central role.Citation3–5) We recently discovered that R. solanacearum strains possess a phc QS system mediated by either (R)-methyl 3-hydroxymyristate or (R)-methyl 3-hydroxypalmitate.Citation6) In addition to EPS, phc QS also regulates the production of secondary metabolites, and ralfuranones A (1) and B (2) (Fig. ) were identified as phc QS-dependent secondary metabolites from R. solanacearum strain GMI1000, a widely used model strain.Citation7) Thereafter, we reported the identification of three new derivatives, ralfuranones J (3), K (4), and L (5) (Fig. ), from R. solanacearum strain OE1-1, a strain isolated from egg plants in Japan, and the involvement of the production of ralfuranone in its virulence in tomato plants.Citation8) Since ralfuranones did not exhibit apparent toxicity against host plants, these compounds may enhance bacterial activities related to virulence.
Fig. 2. HPLC chromatograms of EtOAc extracts from ∆ralA cultures fed with 2.5 μM (A) and 200 μM ralfuranone I (6) (B) and MGRLS medium fed with 2.5 μM ralfuranone I (6) (C). The peaks of ralfuranones were marked with arrowheads.
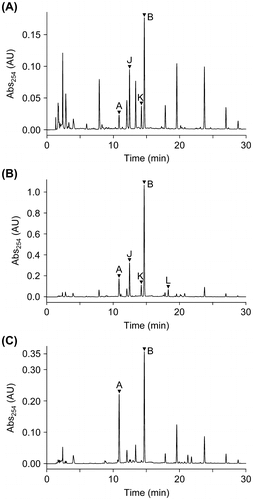
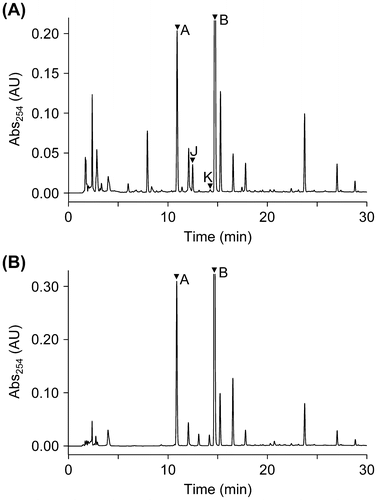
Fig. 3. HPLC chromatograms of EtOAc extracts from ∆ralA cultures (A) and MGRLS medium fed with 10 μM ralfuranone B (2) (B). The peaks of ralfuranones were marked with arrowheads.
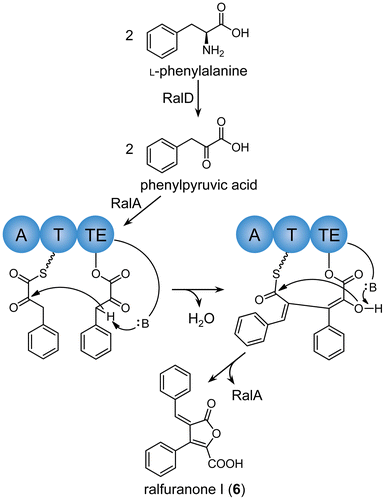
The key enzymes involved in ralfuranone biosynthesis have been studied in detail. The aminotransferase RalD converts l-phenylalanine to phenylpyruvic acid, which is then loaded to the furanone synthase RalA, composed of a tridomain nonribosomal peptide synthetase (NRPS)-like enzyme (Scheme ).Citation9) Two phenylpyruvic acids are condensed by an aldol reaction on RalA, and the product is released from the enzyme by ester hydrolysis. The condensation product, named ralfuranone I (6) (Scheme ), has been suggested as a common biosynthetic precursor for other ralfuranones.Citation10) However, these conversions have not yet been examined in R. solanacearum. We previously reported that a ralA-deletion mutant of OE1-1 (∆ralA) did not produce any ralfuranones, but started to produce them again when the ralA gene was complemented.Citation8) This finding indicated that enzymes involved in the biosynthesis of ralfuranones other than ralA remain active in ∆ralA. Thus, using this mutant, feeding studies on ralfuranones may be conducted without preparing their isotopically labeled precursors. In order to elucidate the biosynthetic pathway of ralfuranones in R. solanacearum, we herein prepared ralfuranone I (6) by culturing OE1-1 and then conducted feeding experiments on ∆ralA. The results obtained showed that the key steps in ralfuranone biosynthesis (ralfuranone I (6) → B (2) → A (1)) proceeded in a non-enzymatic manner.
Experimental
General methods
Ralfuranones A (1), B (2), J (3), K (4), and L (5) were synthesized in a previous study.Citation8) 1H NMR spectra were recorded on a JNM-AL400 spectrometer (JEOL, Tokyo, Japan). Chemical shifts were reported as δ values (ppm); CDCl3 (δH = 7.26) was used as an internal reference. HPLC experiments were performed with a LaChrom Elite HPLC system (Hitachi High-Technologies, Tokyo, Japan) and Prominence HPLC system (Shimadzu, Kyoto, Japan). Solvents for HPLC were purchased from Kanto Chemical (Tokyo, Japan). ESI-MS data were obtained with an LCMS-2020 spectrometer (Shimadzu, Kyoto, Japan).
Bacterial strains and growth conditions
Seed cultures of OE1-1 and ∆ralA were grown in B mediumCitation8) at 30 °C overnight. These were centrifuged to remove medium, and the bacterial cells were then suspended in the same volume of MGRLS medium (MGRL mediumCitation8) supplemented with 3% sucrose). The suspensions were diluted with MGRLS medium in Erlenmeyer flasks and incubated at 30 °C with rotation (130 rpm).
Isolation of ralfuranone I (6)
OE1-1 was grown in MGRLS medium (3 L) containing 10 mM l-phenylalanine and Amberlite XAD-16 adsorption resin (30 g, Sigma, St Louis, USA) at 30 °C with rotation (130 rpm) for 24 h. Following growth, the resin was collected, washed with H2O and 20% MeOH, and eluted with MeOH. The eluate was concentrated and subjected to preparative HPLC. The HPLC conditions were the following: column, Inertsil ODS-P (250 × 10 mm, 5 μm; GL Sciences, Tokyo, Japan); column oven, 40 °C; flow rate, 4 mL/min; eluent, MeCN/H2O (20–95% linear gradient in 40 min, then 95% MeCN for 10 min). Ralfuranone I (6) was obtained as a yellow oil (4 mg). 1H NMR (400 MHz, CDCl3) δ: 7.12 (1H, s, H-10), 7.35 (2H, dd, J = 2.9, 6.8 Hz, H-7), 7.44 (2H, t, J = 7.3 Hz, H-13), 7.49 (2H, m, H-9,14), 7.50 (2H, m, H-8), 8.16 (2H, d, J = 7.3 Hz, H-12). The UV and ESI-MS data of ralfuranone I (6) were shown in Fig. S1.
Feeding of ralfuranones
∆ralA was grown in MGRLS medium (100 mL) at 30 °C with rotation (130 rpm) for 2 days. EtOH solutions of the isolated ralfuranone I (6) or synthetic ralfuranone B (2) were added to the culture, and the bacterial cultures were incubated for a further 2 days. Following growth, the bacterial cultures were extracted three times with an equal volume of EtOAc. The combined EtOAc extracts were dried over Na2SO4 and evaporated to dryness. The residues were dissolved in MeOH (500 μL for experiments with 2.5 μM ralfuranone I (6) and 10 μM B (2), 5 mL for experiments with 200 μM I (6)) and subjected to an HPLC analysis: column, InertSustain C18 (150 × 4.6 mm, 3 μm, GL Sciences); column oven, 40 °C; flow rate, 1 mL/min; eluent, MeCN in H2O (20–95% linear gradient in 24 min, then 95% MeCN for 6 min); injection volume, 10 μL. As control experiments, ralfuranones I (6) or B (2) were added to MGRLS media and incubated at 30 °C with rotation (130 rpm) for 2 days. The analytical samples were prepared in accordance with the method for bacterial samples. Quantification data were shown as the mean ± SD of three independent experiments.
Analysis of benzaldehyde formation
MGRLS media (2 mL) containing 1 mM ralfuranone B (2) were incubated at 30 °C with shaking. After appropriate incubation times (0–100 h), the media (10 μL) were directly analyzed by HPLC.
Results and discussion
We initially determined whether OE1-1 also produced ralfuranone I (6). Due to its instability, ralfuranone I (6), when it diffused into medium, was easily decomposed and, thus, was not recovered from bacterial cultures.Citation7,8,10) In order to collect ralfuranone I (6) effectively, Pauly et al. cultivated GMI1000 in 1/4 M63 medium containing XAD-16 adsorption resin.Citation10) We previously confirmed that OE1-1 produced higher levels of ralfuranones in MGRLS medium than in 1/4 M63 medium.Citation8) Therefore, OE1-1 was grown in MGRLS medium containing XAD-16 resin in this study. After this incubation, the resin was collected, washed with H2O and 20% aq MeOH, and eluted with MeOH. An HPLC analysis of the eluate gave a major peak at tR 16.9 min, which showed a UV spectrum characteristic of ralfuranone I (6) (Fig. S1(A) and (B)).Citation10) Thus, this peak was purified from the eluate of XAD-16 resin incubated in the OE1-1 culture (3 L) by HPLC. The ESI-MS of purified ralfuranone I (6) yielded an [M−H]− ion at m/z 291 and [M−H−CO2]− ion at m/z 247, which corresponded to ions with the molecular weight of 6 (Fig. S1(C)). The 1H NMR data of ralfuranone I (6) were identical to previous findings.Citation10) Thus, we confirmed that OE1-1 also produced ralfuranone I (6).
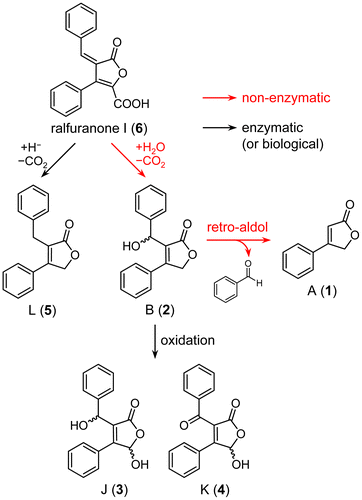
OE1-1 has been shown to produce ralfuranones A (1), B (2), J (3), K (4), and L (5) in MGRLS medium.Citation8) Using purified ralfuranone I (6), we conducted feeding experiments to determine whether ralfuranone I (6) was converted to these ralfuranones in ∆ralA. After the pre-incubation of ∆ralA for 48 h in MGRLS medium, ralfuranone I (6) was added to the bacterial culture and was then incubated for a further 48 h. As a control, ralfuranone I (6) was incubated for 48 h in MGRLS medium without ∆ralA. These cultures were extracted with EtOAc, and the extracts were analyzed by HPLC. Ralfuranones A (1), B (2), J (3), and K (4) were detected in extracts of the ∆ralA culture fed with 2.5 μM ralfuranone I (6) (Fig. (A)). Based on standard curves, we quantified the concentrations of ralfuranones as A (1) 59 ± 4, B (2) 837 ± 12, J (3) 331 ± 42, and K (4) 68 ± 9 nM. The production of ralfuranone L (5) (2.43 ± 0.36 μM) was observed when ∆ralA was grown in medium containing 200 μM ralfuranone I (6) (Fig. (B)). Other ralfuranones were detected as follows: A (1) 5.01 ± 0.77, B (2) 59.2 ± 6.5, J (3) 14.1 ± 1.54, and K (4) 2.51 ± 0.77 μM. However, the conversion of ralfuranone I (6) to ralfuranones A (1) (612 ± 143 nM) and B (2) (1870 ± 400 nM) was also observed in MGRLS medium (containing 2.5 μM ralfuranone I (6)) without ∆ralA (Fig. (C)), indicating that these conversions were non-enzymatic. Since the significant formation of ralfuranones J (3), K (4), and L (5) was not observed in this extract, these compounds may depend on the activity of R. solanacearum cells. In these conversion experiments, the level of ralfuranone B (2) was higher than that of ralfuranone A (1), implying the following conversion pathway: ralfuranone I (6) → B (2) → A (1).
We previously fed deuterium-labeled ralfuranone B (2) to the OE1-1 culture and observed that it was metabolized to ralfuranones J (3) and K (4).Citation8) However, the deuterium labels in the upper benzene ring of ralfuranone B (2) were missing when ralfuranone B (2) was converted to A (1). Furthermore, the above feeding experiments with ralfuranone I (6) suggested that the conversion of ralfuranone B (2) to A (1) may be non-enzymatic. In order to clarify this, feeding experiments with ralfuranone B (2) were performed at a concentration of 10 μM in MGRLS medium with and without ∆ralA. An HPLC analysis of the extract of the ∆ralA culture gave peaks corresponding to ralfuranones A (1), J (3), and K (4) (Fig. (A)). The level of ralfuranone A (1) (415 ± 120 nM) was higher than those of ralfuranones J (3) (127 ± 14 nM) and K (4) (8 ± 1 nM). The extract of the MGRLS medium without ∆ralA also gave ralfuranone A (1) (892 ± 24 nM), but not J (3) or K (4) (Fig. (B)). Thus, we confirmed that ralfuranone B (2) was non-enzymatically converted to ralfuranone A (1), but enzymatically (or biologically) to ralfuranones J (3) and K (4).
The results of the feeding and conversion experiments suggested a pathway for the biosynthesis of ralfuranones in R. solanacearum, as shown in Scheme . Ralfuranone B (2) was identified as the main product of ralfuranone I (6). This conversion may have occurred non-enzymatically through the attack of H2O on the exocyclic double bond and subsequent decarboxylation and was also supported by the absolute configuration of the oxymethine at C-10 in ralfuranone B (2) being racemic.Citation8) Furthermore, Pauly et al. reported that ralfuranone I (6) non-enzymatically reacted with thiols by the Michael reaction, and the products underwent decarboxylation to form various artificial ralfuranones.Citation10) This reaction manner may essentially be the same as the formation of ralfuranone B (2) from I (6). The conversion of ralfuranone B (2) to A (1) may have spontaneously occurred through a retro-aldol reaction. When ralfuranone B (2) was incubated in MGRLS medium, benzaldehyde accumulated in the medium in accordance with the formation of ralfuranone A (1) (Fig. S2). This result suggests the involvement of a retro-aldol reaction in the conversion of ralfuranone B (2) to A (1). To the best of our knowledge, this is the first time that such a unique reaction has led to the formation of natural furanones. As described previously,Citation8) ralfuranones J (3) and K (4) were the enzymatic oxidation products of ralfuranone B (2). We expected ralfuranone B (2) to be oxidized to J (3) and then further oxidized to K (4) in OE1-1; however, the reaction order and enzymes involved were not examined in this study. Ralfuranone L (5) was synthesized from ralfuranone I (6) presumably by the attack of hydride on the exocyclic double bond and subsequent decarboxylation. Since it may be hard to form hydride in medium, we expected this conversion to need R. solanacearum cells and an enzyme.
In conclusion, we herein demonstrated that ralfuranone I (6) was a precursor for other ralfuranones in R. solanacearum. By analyzing the conversion of ralfuranones I (6) and B (2), we suggested a biosynthetic pathway for ralfuranones, in which R. solanacearum uniquely synthesized the aryl-type furanones by combining non-enzymatic and enzymatic reactions. In a previous study, ∆ralA exhibited reduced virulence in host plants as well as the ablation of ralfuranone production.Citation8) These findings strongly suggest that ralfuranones play an important role in this wilt disease. We are now investigating the mechanisms by which ralfuranones enhance bacterial activity in order to induce wilt disease in host plants. The present results may indicate that ralfuranone I (6) does not exhibit important biological activity due to its rapid conversion to other ralfuranones. Future studies on these unique metabolites may provide an insight into the ability of R. solanacearum to cause wilting in host plants.
Author contributions
K.K. designed the experiments. K.K. and H.O. performed the experiments. K.K., H.O, A.K. K.O., and Y.H. analyzed the data. K.K., H.O., and Y.H. contributed reagents and materials. K.K. and Y.H. wrote the paper.
Funding
This work was supported by the Agricultural Chemical Research Foundation; Japan Society for the Promotion of Science [grant number 25292029], [grant number 26660036]; Osaka Prefecture University a grant for young scientists.
Supplemental materials
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2015.1116931.
SOM151014.pdf
Download PDF (214 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Hayward HC. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 1991;29:65–87.
- Schell MA. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 2000;38:263–292.
- Genin S, Denny TP. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 2012;50:67–89.
- Flavier AB, Clough SJ, Schell MA, et al. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 1997;26:251–259.
- Clough SJ, Lee KE, Schell MA, et al. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxyplamitic acid methyl ester. J. Bacteriol. 1997;179:3639–3648.
- Kai K, Ohnishi H, Shimatani M, et al. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. ChemBioChem. 2015;16:2309–2318.
- Schneider P, Jacobs JM, Neres J, et al. The global virulence regulators VsrAD and PhcA control secondary metabolism in the plant pathogen Ralstonia solanacearum. ChemBioChem. 2009;10:2730–2732.
- Kai K, Ohnishi H, Mori Y, et al. Involvement of ralfuranone production in the virulence of Ralstonia solanacearum OE1-1. ChemBioChem. 2014;15:2590–2597.
- Wackler B, Schneider P, Jacobs JM, et al. Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem. Biol. 2011;18:354–360.
- Pauly J, Spiteller D, Linz J, et al. Ralfuranone thioether production by the plant pathogen Ralstonia solanacearum. ChemBioChem. 2013;14:2169–2178.