Abstract
Lactation is a common feeding strategy of eutherian mammals, but its functions go beyond feeding the neonates. Ever since Tissier isolated bifidobacteria from the stool of breast-fed infants, human milk has been postulated to contain compounds that selectively stimulate the growth of bifidobacteria in intestines. However, until relatively recently, there have been no reports to link human milk compound(s) with bifidobacterial physiology. Over the past decade, successive studies have demonstrated that infant-gut-associated bifidobacteria are equipped with genetic and enzymatic toolsets dedicated to assimilation of host-derived glycans, especially human milk oligosaccharides (HMOs). Among gut microbes, the presence of enzymes required for degrading HMOs with type-1 chains is essentially limited to infant-gut-associated bifidobacteria, suggesting HMOs serve as selected nutrients for the bacteria. In this study, I shortly discuss the research on bifidobacteria and HMOs from a historical perspective and summarize the roles of bifidobacterial enzymes in the assimilation of HMOs with type-1 chains. Based on this overview, I suggest the co-evolution between bifidobacteria and human beings mediated by HMOs.
Graphical abstract
Symbiosis and co-evolution between bifidobacteria and humans, mediated by human milk oligosaccharides with a type-1 chain.
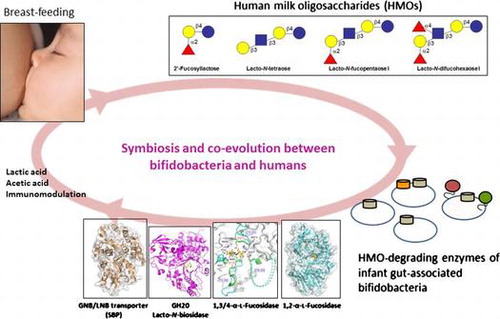
I. Preface
The mammalian gastrointestinal tract is inhabited by a vast and diverse community of microbesCitation1) and their metabolic activity significantly influences the host’s health. In mice, short-chain fatty acids produced by intestinal bacteria have been shown to affect the body weight, energy balance, and immune response,Citation2) while butyric acid produced by clostridia is known to induce the differentiation of colonic regulatory T cells and also induce the expression of N-acetylglucosamine 6-O-sulfotransferase-2 that modifies mucin-type O-glycans in the gut.Citation3,4) Moreover, the presence of certain bacterial metabolites has frequently been associated with the incidence of behavioral abnormalities of the host.Citation5) It is therefore not exaggerated to consider the intestinal bacterial population as an organ of the host. Hence, it is important to understand what shapes and influences this consortium of intestinal bacteria.
Dietary habits have been assumed to significantly affect the gut microbial composition. This concept is true in most part, because gut microbiomes are shown to vary among communities with different dietsCitation6) and also to change rapidly within individuals when their diets change.Citation7) Non-digestible glycans in diets are thus the main factor affecting the gut microbial community. The genomes of several gut microbes contain genetic loci responsible for the utilization of plant-derived glycans, such as pectic polysaccharides and xyloglucans, and the foraging activity of these bacteria allows them to thrive in the gut ecosystem.Citation8) Our group reported the isolation of the genes encoding 1,2-α-L-fucosidase and endo-α-N-acetylgalactosaminidase from bifidobacteria in 2004 and 2005, respectively.Citation9,10) Both enzymes act on host-derived glycans, including milk oligosaccharides and mucin glycoproteins, and interestingly, the homologs of these enzymes were found to be conserved in particular gut microbes.Citation11) Based on these findings, we envisaged that the host-derived glycans are also an important and influential factor for the microbial composition in the gutCitation11) (Note that a host genetic factor, e.g., secretor or non-secretor, has been recently shown to affect gut microbiota composition.Citation12))
The composition of human gut microbiota changes during life, and the most drastic adaptations occur at the start of breast-feeding and the end of it, i.e., during weaning.Citation13,14) The intestines of breast-fed infants are rapidly and dominantly colonized by bifidobacteria within a week after birth, and the bifidobacteria-rich population drastically decreases after weaning.Citation15,16) This observation strongly suggests that human milk contains compounds (perhaps, host-derived glycans) that selectively stimulate the growth of bifidobacteria in the infant gut. The presence of bifidobacteria in the gut microbiota appears to be correlated with a reduced incidence of diseases such as diarrhea and allergies in infants.Citation17) Various beneficial effects of bifidobacteria on host health have been reported, including the inhibition of harmful bacteria,Citation17) fortification of barrier function of intestinal epithelial cells,Citation18) and modulation of immune function.Citation19) Thus, the bacteria might be the first evolutionary commensals in the human gut, but the molecular mechanism underlying the formation of bifidobacteria-rich microbiota in the intestines of breast-fed infants remains elusive. In this review, I summarize the advances made by our group over the past 10 years on bifidobacterial enzymes that are dedicated to the degradation of human milk oligosaccharides (HMOs; see section III for definition), in the context of the historical research efforts on bifidobacteria and HMOs. The structure–function analysis of the bifidobacterial enzymes indicates a symbiosis and co-evolution between bifidobacteria and humans.
II. Historical background
Bifidobacterium sp. (first named Bacillus bifidus communis) was isolated in 1899 from the feces of breast-fed infants by H. Tissier at the Pasteur Institute. He already mentioned at the time that the gut microbiota of breast-fed infants are richer in bifidobacteria as compared to the feces of bottle-fed infants.Citation20) Although the ratio of occupancy varies in different studies and with different detection techniques (culture method vs. non-culture method), it is currently well established that an increased population of bifidobacteria and a decreased population of other, especially pathogenic, bacteria, are specific features of the intestinal microflora of breast-fed babies.Citation13–15) The finding that bifidobacteria are predominantly present in the gut of breast-fed newborns and that the bacteria showed enhanced growth on media supplemented with human, but not cow, milk,Citation20,21) convinced researchers that human milk contains bifidogenic compounds (or bifidus factors).
In 1954, Gauhe et al. reported that the oligosaccharides in human milk have a bifidogenic effect.Citation22) They partially purified the relevant oligosaccharides and found they consisted of L-fucose (Fuc), galactose (Gal), glucose (Glc), and N-acetylglucosamine (GlcNAc). The concept that HMOs acted as bifidus factors was attractive. However, the interpretation of these experiments was later disputed, because they used a variant strain of Bifidobacterium sp. (Bifidobacterium bifidum var. pennsylvanicus) that was probably impaired in its ability to synthesize GlcNAc-containing cell wall components.Citation22,23) It should be mentioned, however, that most bifidobacteria do not utilize exogenously added GlcNAc efficiently.Citation24) Therefore, the above observation should not be attributed to misuse of the variant strain only. Indeed, Gauhe et al. mentioned in their study that many bifidobacterial strains isolated from the breast-fed infants showed the same phenotype as Bifidobacterium bifidum var. pennsylvanicus.Citation21) Since then and up to 2004, there were no reports indicating a biological connection between HMOs and bifidobacteria. In the last two decades, many researchers focused instead on the anti-microbial and immunomodulatory activities of HMOs. Ruiz-Palacios et al. showed that HMOs with an H-antigen structure (Fucα1-2Gal) inhibit Campylobacter jejuni infection in an in vivo mouse model,Citation25) and Crane et al. found that HMOs block the binding of enterotoxins of Escherichia coli to intestinal cells.Citation26) Reduction of the binding of human immunodeficiency virus-1 to dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin was also reported.Citation27) In addition, disialyllacto-N-tetraose was shown to ameliorate necrotizing enterocolitis in rats.Citation28) These studies, combined with a continuous advance of analytical instruments, have become the driving force to decipher various biological functions of HMOs.
III. Human milk oligosaccharides (HMOs)
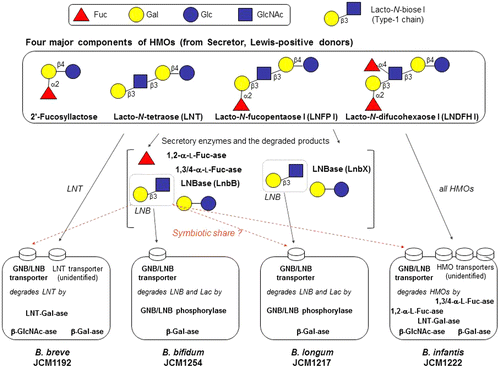
Oligosaccharides (with a degree of polymerization of more than 3) are the third most abundant solid component in human milk (between 10 and 20 g/L), after lactose (Lac) and lipids, and they are collectively termed HMOs.Citation29) HMOs are characterized by their complex structures, and more than 200 molecular species have been detected.Citation30) They are divided into 13 core structures that consist of Lac at the reducing end, elongated by β1-3-linked lacto-N-biose I (Galβ1-3GlcNAc: LNB, type-1 chain) and/or β1-3/6-linked N-acetyllactosamine (Galβ1-4GlcNAc: LacNAc, type-2 chain). The core structures are frequently modified by Fuc and sialic acid (Neu5Ac) via α1-2/3/4 and α2-3/6 linkages, respectively, to more complex structures.Citation31) Among these HMOs, lacto-N-tetraose (LNT, Galβ1-3GlcNAcβ1-3Galβ1-4Glc), lacto-N-fucopentaose I (LNFP I, Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc), lacto-N-difucohexaose I (LNDFH I, Fucα1-2Galβ1-3(Fucα1-4)GlcNAcβ1-3Galβ1-4Glc), and 2′-fucosyllactose (2′-FL, Fucα1-2Galβ1-4Glc) are the most abundant, especially in the colostrum, unless the milk is derived from non-secretor and/or Lewis-negative subjects (Fig. ).Citation29,31) It should be noted that HMOs are especially rich in type-1 chains, as is the case for three of the above structures (LNT, LNFP I, and LNDFH I), as this composition is unique for human milk oligosaccharides.Citation29) The type-1 richness of HMOs constitutes one of the rationales behind the concept of symbiosis and co-evolution between bifidobacteria and humans, details of which are described in sections IV and V.
Fig. 1. The structure of four main HMOs, and the enzymes involved in the degradation of the HMOs in infant-gut-associated bifidobacteria.
Considering the mother’s energy expenditure to synthesize 10–20 g/L of HMOs in the mammary glands (one ATP is theoretically consumed per elongation of one sugar residue), they should represent a relevant biological advantage to infants.Citation32) One important activity of HMOs is, as mentioned above, to defend neonates from infectious disease. HMOs are resistant to the host’s intestinal digestive enzymes,Citation33) and thus the majority should reach the colon, where they exert their activity. Another activity of HMOs is the modulation of immune responses.Citation17,19) A small amount of HMOs is detected in the urine and plasma of infants, sometimes modified by acetylation and further glycosylation.Citation34,35) He et al. showed that 2′-FL can attenuate the lipopolysaccharide-induced inflammatory response of intestinal epithelial cells in vitro.Citation36) As discussed in this review, one of the most important activities of HMOs is to enhance the development of a bifidobacteria-rich microbial population, by acting as their preferred nutrient. Infant-gut-associated bifidobacteria possess the specific genetic and enzymatic toolsets dedicated to the assimilation of HMOs.
IV. Specific occurrence of HMO-degrading enzymes in infant-gut-associated bifidobacteria
Bifidobacterium breve, B. bifidum, B. longum subsp. infantis (referred to as B. infantis), and B. longum subsp. longum (B. longum) are four well-known species/subspecies found in the stool of breast-fed babies.Citation15,37) In 1999, Derensy-Dron et al. reported the presence of a phosphorylase specific for galacto-N-biose (GNB, Galβ1-3GalNAc) and lacto-N-biose I (LNB, Galβ1-3GlcNAc) in B. bifidum.Citation38) The enzyme catalyzes the phosphorolysis of GNB and LNB to produce α-D-galactose 1-phosphate and the respective N-acetylhexosamines. Subsequently, Kitaoka et al. isolated the corresponding gene from B. longum and fully characterized the enzyme.Citation39) GNB is the disaccharide liberated from core-1 type O-glycan (Galβ1-3GalNAcα-O-Ser/Thr) by endo-α-N-acetylgalactosaminidase (isolated from B. longum) (see Section I).Citation10) Given that GNB/LNB phosphorylase (GLNBP) is located in the cytoplasm while endo-α-N-acetylgalactosaminidase is cell wall anchored with its catalytic domain exposed to the outer surface, the liberated GNB needs being imported by a specific transporter. Not surprisingly, the genes encoding an unidentified ATP-binding cassette (ABC)-type sugar transporter were found upstream of the GLNBP gene. The presence of GLNBP (intracellular) and endo-α-N-acetylgalactosaminidase (extracellular) in bifidobacteria also suggested that these organisms possess an extracellular enzyme that liberates LNB from host-derived glycans. LNB (type-1 chain) is present as a constituent of glycan chains of various glycoconjugates, including glycosphingolipids, mucin glycoproteins, and importantly HMOs (Fig. . see Section III).Citation31) Kitaoka et al. thus proposed that LNB is a bifidus factor.Citation39) Accordingly, the molecular cloning of 1,2-α-L-fucosidase, endo-α-N-acetylgalactosaminidase, and GLNBP became an important clue to elucidate the disputed role of HMOs as bifidus factors. In the following sections, I shortly describe the key enzymes required to degrade the three main components of type-1 HMOs (LNT, LNFP I, and LNDFH I) (Fig. ).
Lacto-N-biosidase (LNBase)
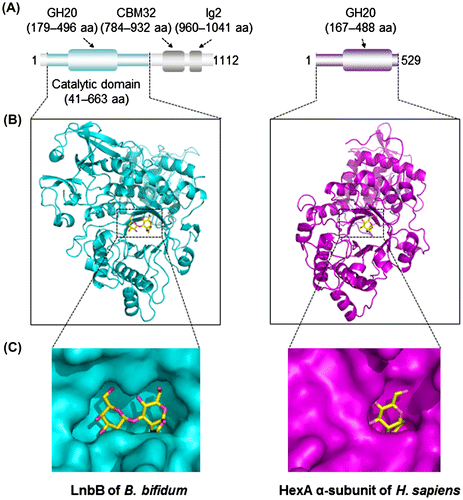
Twenty-eight bacterial species/subspecies belonging to the Bacteroides, Bifidobacterium, Clostridium, Enterococcus, Eubacterium, Lactobacillus, and Ruminococcus genera, which constitute main human gut microbiome, were examined for their ability to liberate LNB from LNT.Citation40) Of these 28 (sub)species, all strains of B. bifidum and two strains of B. longum tested positive (LNBase+). The gene for LNBase was isolated from B. bifidum based on the sequence of the Streptomyces sp. LNBase, which was the sole enzyme identified at that time.Citation41) The gene, designated lnbB, was found to encode a protein of 1,112 amino acid residues with a predicted molecular mass of 120 kDa. The deduced amino acid sequence contained a signal peptide and a membrane anchor at the N-terminal (1–34 aa) and C-terminal (1082–1108 aa) ends. The database search revealed the presence of an N-terminal glycoside hydrolase family 20 (GH20) domain (179–496 aa),Citation42) a central carbohydrate-binding module 32 (CBM32; 784–932 aa), and a C-terminal bacterial immunoglobulin (Ig)-like 2 domain (960–1041 aa)Citation40) (Fig. (A)). The LnbB protein exhibited 38% amino acid identity to the LNBase from Streptomyces sp. and 20 to 25% identity to β-N-acetylhexosaminidases (β-HexNAc-ases) from bacterial and mammalian sources.Citation43,44) Hydrolysis by GH20 enzymes is known to proceed through a substrate-assisted mechanism, in which the N-acetyl group of the substrate (in the case of LNT, this is GlcNAc) attacks its own anomeric carbon to form an oxazoline intermediate.Citation44) The important catalytic residues identified in β-Hex-ases were conserved in LnbB.Citation40) Interestingly, while LNBase acts on β-linked LNB and to a lesser extent on β-linked GNB, it does not act on β-linked HexNAc monosaccharides. In contrast, β-HexNAc-ases act on β-linked HexNAc, but not on β-linked LNB/GNB disaccharides, even though the linkage to be hydrolyzed is the same and both enzymes belong to the same GH family.Citation40,43–45) To understand the molecular basis of the substrate recognition by each enzyme and to subsequently be able to distinguish between the two at the sequence level, our group solved the crystal structure of the catalytic domain of LnbB (41–663 aa) in complex with LNB at 1.8-Å resolution (Fig. (B)).Citation46) The structures of complexes with competitive inhibitors with Ki values of 0.1–50 μM have recently also been determined.Citation47) As shown in the molecular surface models of the two enzymes (B. bifidum LnbB and human β-HexNAc-ase),Citation45) LNBase possesses an extended pocket (-2 subsite) suitable for accommodating the β-(1→3)-linked Gal residue of LNB (Fig. (C)) and forms several hydrogen bonds with Gal.Citation46) In contrast to LNBase, Hsβ-HexNAc-ase possesses a catalytic pocket that can accept one HexNAc residue and the corresponding site (Gal-binding site of LNBase) is occupied by the residues extending from the β1 and β2 sheets and the following loop region, which become a steric hindrance for the accommodation of LNB. LNBase lacks a residue that recognizes the O3 atom of −1 GlcNAc, which could prevent it from forming a stable transition-state intermediate in the absence of the −2 Gal residue, leading to the loss of HexNAc-ase activity. As this information provided us with a rationale for how the two enzymes differently recognize their respective substrates, we were able to discriminate between the two enzymes at the amino acid sequence level. The homologs of LnbB (i.e., LNBase, not β-HexNAc-ase) are found in the genomes of B. bifidum (all of the sequenced strains), Trueperella pyogenes (an isolate from bovine mastitis), Actinomyces neuii (an isolate from mammary prosthesis), Chlamydia trachomatis, and some species of Streptomyces, which indicates the specific presence of LNBase in bifidobacteria. The structural studies also suggested that LNBase evolved from β-HexNAc-ase.
Fig. 2. Structural differences between B. bifidum GH20 lacto-N-biosidase (LnbB) and human GH20 N-acetylhexosaminidase (HexA).
The genome sequence of B. longum JCM1217 became publicly available in 2011.Citation48) Although the strain was shown to be phenotypically LNBase+, no homologous gene to lnbB of B. bifidum was found. The gene product containing the GH20 domain was identified to be β-GlcNAc-ase.Citation49) Expression cloning was carried out to isolate the LNBase gene from the strain, and consequently, locus_tag BLLJ_1505 and BLLJ_1506 were identified.Citation50) BLLJ_1505 (termed LnbX) showed no sequence similarity to the previously characterized proteins but was found to possess a signal peptide (1–30 aa), a right-handed β-helix region (158–315 aa), an uncharacterized sugar-binding domain called FIVAR (1435–1487 aa), and a membrane anchor (1577–1593 aa). BLLJ_1506 (termed LnbY) has a signal peptide (1–29 aa) as well, but lacks any functional motifs. Enzymatic and genetic characterization revealed that LNBase from B. longum is comprised of the gene product of lnbX only, while the gene product of lnbY acts as a designated chaperon for LnbX. The purified LnbX contained Ca2+ and Mg2+ ions, and denatured LnbX efficiently refolded when both LnbY and the two metal ions were present. LnbY did not contain any metal ion. The lnbX and lnbY genes constitute an operon, as the polar effect was observed when the lnbX gene was disrupted in B. longum.Citation50) The substrate specificity of LnbX was also found to be unique. Although GH20 LnbB is active on the unmodified LNB structure (e.g., LNT), LnbX was able to hydrolyze LNFP I and sialyllacto-N-tetraose a (LST a) to release 2′-fucosyl LNB and 3′-sialyl LNB, respectively. However, the activities are considerably lower than that for LNT, and therefore not physiologically relevant (Table . See section V). Moreover, LnbX was found to act on the β-linked GNB present in sugar chains of globosides (Gb5 and globo H), but it was inactive on GA1 tetrasaccharide in which β-GNB is linked with the axial O4 of the Gal residue. GH20 LnbB was capable of acting on GA1 and Gb5 oligosaccharides but not on globo H hexasaccharide.Citation51) LnbX homologs are found in the genomes of gut microbes belonging to Bifidobacterium, Roseburia, Eubacterium, Ruminococcus, and Clostridium, whereas LnbY homologs are found only in B. longum and B. bifidum (note that LnbY homologs of B. bifidum lack a signal sequence).Citation52) Considering the unique maturation process of LnbX and LnbY, it should be empirically determined whether these homologs have LNBase activity. Structural studies of LnbX and LnbY are currently ongoing, which will help us understand the unique features of these two enzymes.
Table 1. Substrate specificities of type-1 chain-specific glycosidases from infant-gut-associated bifidobacteria.
Galacto-N-biose/lacto-N-biose I transporter (GNB/LNB transporter)
As mentioned above, the genes for an ABC transporter were found to be located just upstream of the GNB/LNB phosphorylase (GLNBP) gene in the B. longum genome.Citation39) When they were introduced into B. longum strain 105-A on a plasmid, the resulting strain was found to show a 10-fold increase in the uptake of (14C-labeled) LNB as compared with the control strain carrying an empty plasmid (unpublished results). This indicates the direct involvement of the genes in LNB transport. The substrate specificity of the ABC transporter is primarily defined by the solute-binding protein, and therefore we purified and characterized the corresponding protein of the transporter.Citation53) The purified protein (termed GL-BP) was shown by isothermal calorimeter analysis to specifically bind GNB and LNB with a Kd value of 10 and 87 nM, respectively. The protein had a low affinity for LNT (Kd of 11 μM), but it was completely heat silent toward monosaccharides constituting the disaccharides (i.e., Gal, GlcNAc, and GalNAc), and disaccharides Lac and LacNAc (type-2 chain). The binding was enthalpy-driven with a slight negative entropy change. Subsequent X-ray structural analysis revealed that the binding was primarily dependent on hydrogen bonds. Interestingly, the axial O4 of the GalNAc residue of GNB forms a water-mediated hydrogen bond with the GL-BP protein, which is absent in the LNB complex. This might correspond to the difference in enthalpy change in the binding of the two ligands (8 kcal/mol).Citation53) It should be noted that the genomic locus of this transporter in B. longum lacks a gene encoding an ATP-binding protein, required to provide energy for the transport. However, it is not rare for gram-positive bacteria to use one ATP-binding protein to energize several ABC transporters.Citation54) Homologs of the LNB/GNB transporter were found in the genomes of several bifidobacteria (B. longum, B. bifidum, B. breve, B. infantis, B. pseudocatenulatum, B. animalis, and B. pseudolongum), as well as in the genomes of skin-, and vagina-colonizing bacteria such as Propionibacterium acnes and Mobiluncus curtisii, respectively. Thus, the distribution of the LNB/GNB transporter in gut microbes is exclusively limited to bifidobacteria, and to the best of our knowledge, all strains belonging to infant-gut-associated bifidobacteria (B. longum, B. bifidum, B. breve, B. infantis) possess the gene cluster. GNB/LNB phosphorylase (GLNBP), isolated by Kitaoka et al., is also exclusively found in the genomes of bifidobacteria.Citation39)
Lacto-N-tetraose β-1,3-galactosidase (LNT-Gal-ase)
In 2008, Sela et al. determined the genomic sequence of B. infantis.Citation55) They found that this subspecies has a gene cluster dedicated to the degradation of HMOs (termed HMO cluster-1) on its genome and hypothesized that the presence of this cluster is linked to the ability of the respective strains to grow on HMOs. Interestingly, none of the glycosidases encoded in the cluster were found to possess a signal sequence. Therefore, Sela et al. predicted that the organism imports intact HMOs into the cytoplasm prior to degradation.Citation55) Indeed, when B. infantis cells were incubated with LNT, the tetrasaccharide disappeared from the culture supernatant, and during the incubation period, no mono-, di-, and trisaccharides appeared in the spent medium.Citation56) The uptake involves an ABC transporter, as the activity was completely abolished in the presence of an ABC transporter inhibitor. Degradation of LNT by B. infantis inside the cells involves monosaccharide-releasing exo-glycosidases because liberation of Gal, GlcNAc, and Glc was observed when LNT was incubated with the cell-free extract. This was an intriguing finding because the β-galactosidase (β-Gal-ase) located in the HMO cluster-1 was considered to be specific for type-2 chains as it belongs to GH2.Citation42) Taking these findings into consideration, the genome of this subspecies was scrutinized, and 6 candidate genes were isolated and the recombinant proteins were characterized enzymatically. From these 6 candidates, the protein with locus_tag Blon_2016 (belonging to GH42), not located at the locus HMO cluster-1, was found to efficiently liberate Gal from LNT (kcat/Km of 120 mM−1 s−1) (Table ), while the other enzymes, including GH2 β-Gal-ase from the HMO cluster-1, were inactive on LNT.Citation56) This was the second report at the time describing the isolation of a β-Gal-ase that hydrolyzes type-1 chains, and the first report to show that the GH42 enzyme degrades human-derived glycans. GH42 members had been assumed to degrade plant-derived glycans.Citation42) Interestingly, LNT-Gal-ase was shown to have a higher affinity for LNT than for LNB, even though the scissile bond is the same in the two substrates. Probably, the enzyme has a (+) subsite that interacts with the Lac moiety of LNT. Our results also revealed that the HMO degradation capacity of B. infantis could not be exclusively attributed to HMO cluster-1.Citation56)
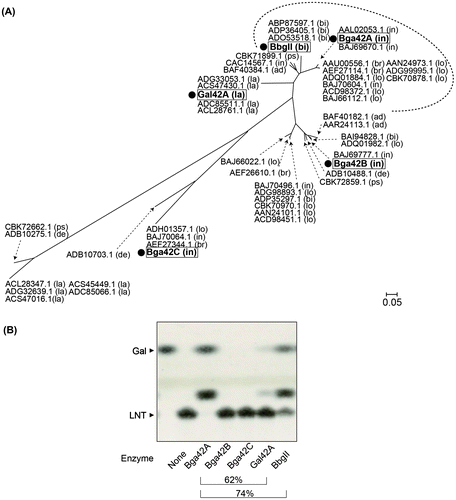
Recent reports by Viborg et al. have shown that the substrate specificity of different GH42 enzymes is more diverse than previously anticipated based on their phylogenetic distances (Fig. (A)).Citation57,58) For example, the amino acid sequence of Gal-ase (locus_tag Balac_0484, termed BlGal42A) from B. animalis subsp. lactis is 62% identical to that of LNT-Gal-ase of B. infantis. Nonetheless, it hardly acts on LNT (Fig. (B)) but shows a preference for galactooligosaccharides with β-(1→6/3) linkages.Citation57) On the other hand, B. bifidum β-Gal-ase, which is 74% identical to B. infantis LNT-Gal-ase, was found to be capable of hydrolyzing LNT although its efficiency is less than that of the B. infantis enzyme (Fig. (B)). Structural studies are necessary to understand the molecular basis of the differential substrate recognition by these similar enzymes at the (+) subsites. From bacteria to humans, only four enzymes (all GH35 and GH42 members) have been reported so far to act on type-1 chains, suggesting that this type of activity is quite rare in nature.Citation42) Homologs (>74% identity) of LNT-Gal-ase of B. infantis were found exclusively in infant-gut-associated bifidobacteria (B. longum, B. bifidum, B. breve, and B. infantis) (Fig. (A)).
Fig. 3. Different substrate specificities of the GH42 members.
Our group also enzymatically and structurally characterized 1,2-α-L-fucosidaseCitation9,59,60) and 1,3-1,4-α-L-fucosidaseCitation61,62) of bifidobacteria. Both enzymes are indispensable for the removal of Fuc residues from HMOs, prior to the action of the three type-1 specific enzymes mentioned above. Lack of α-L-fucosidase activity is known to severely impair the ability of bifidobacteria to utilize HMOs, because about 70% of all HMOs are fucosylated (except for non-secretors).Citation29) GH95 or 1,2-α-L-fucosidase is present in B. bifidum (secretory form), B. infantis (cytosolic form), some strains of B. breve (cytosolic form), and a few gut bacteria belonging to Bacteroides, Clostridium and Ruminococcus, but it has not been identified in B. longum. The other fucosidase, 1,3-1,4-α-L-fucosidase (GH29 subfamily B) removes Fuc residues from Lewis a/x trisaccharide structures (Galβ1-3/4(Fucα1-4/3)GlcNAcβ) but not from Fucα1-4/3GlcNAc disaccharides, and it is found in B. bifidum (secretory form), B. infantis (cytosolic form), and Bacteroides thetaiotaomicron, but not in B. breve and B. longum. The unique feature of 1,3-1,4-α-L-fucosidase that requires a branched Gal residues has been identified by X-ray structural studies.Citation62,63) Lactobacillus casei has a cytoplasmic α-L-fucosidase specific for Fucα1-3GlcNAc disaccharides.Citation64) However, it remains to be elucidated whether this enzyme is involved in HMO degradation because action of this enzyme on HMO structures requires prior removal of Gal residues.
Taken together, these findings suggest that infant-gut-associated bifidobacteria have evolved specific enzymes dedicated to the degradation of type-1 HMOs. The metabolic process of LNB inside the cells has been summarized by Kitaoka.Citation65) The limited occurrence of the homologs of HMO-degrading enzymes in bifidobacteria has also been reported by Odamaki et al.Citation66)
V. Consumption of HMOs by infant-gut-associated bifidobacteria
The ability of certain species of bifidobacteria to assimilate HMOs was first demonstrated by Ward et al. and LoCascio et al. in 2007,Citation67,68) and not much later also by Marcobal et al.Citation69) By analyzing the oligosaccharides present in the spent media by mass spectrometry, these authors found that B. infantis can consume a range of HMO structures, whereas B. breve and B. longum exhibit only a very limited utilization of HMOs. The authors also showed that some Bacteroides sp. degrade HMOs.
Our group first examined LNT (type-1 chain) degradation by various gut microbes and found that this activity is limited to infant-gut-associated bifidobacteria (data not shown). No degradation of LNT, or even liberation of Gal, was detected in the cell suspensions of other intestinal bacteria belonging to Bacteroides, Clostridium, Lactobacillus, Enterococcus, and Eubacterium. Only in the supernatant of B. thetaiotaomicron cultures, a small amount of the trisaccharide lacto-N-triose II (GlcNAcβ1-3Galβ1-4Glc) was identified. Given these results, we examined the in vitro fermentation ability of infant-gut-associated bifidobacteria and precisely determined the metabolic fate of each of the neutral HMOs during cultivation, using HPLC to detect 2-anthranilic-acid-labeled sugars.Citation24) The merits of HPLC detection of fluorescein-labeled sugars over mass spectrometry are (1) highly sensitive quantification of sugars (mono- and oligosaccharides), and (2) reliable isomer separation (type-1 versus type-2). When the selected strains of infant-gut-associated bifidobacteria were incubated in medium containing HMOs as the carbon source, B. bifidum JCM1254 and B. infantis JCM1222 showed vigorous growth (OD600 > 1.5). The growth of B. breve JCM1192 and B. longum JCM1217 was significantly lower, as the OD600 value never exceeded 0.4. HPLC analysis of the mono- and oligosaccharides in the spent media revealed that B. breve JCM1192 and B. longum JCM1217 consumed LNT only (Fig. ). B. longum used lacto-N-biosidase (LnbX) to degrade LNT, as a transient increase in Lac accompanied the decrease in LNT. A slight decrease in LNFP I was observed, which reflects the substrate specificity of LnbX, but this decrease seemed not physiologically relevant. LNB was not detected during this period, suggesting that B. longum prefers LNB over Lac. In contrast, B. breve appeared to consume LNT without hydrolyzing it outside the cells. In other words, B. breve directly imported the tetrasaccharide, because no mono- or di-saccharide degradation products were detected. In accordance, neither LNT-Gal-ase nor LNBase activity was detected in the extracellular fraction of B. breve JCM11192. The genomes of the 7 strains of this species do not possess homologs for the secretory β-galactosidase or for LNBase.Citation52)
Fig. 4. Metabolic fates of the four main HMO components (green: 2′-FL; purple: LNT; cyan: LNFP I; and orange: LNDFH I) during cultivation with infant-gut-associated bifidobacteria.
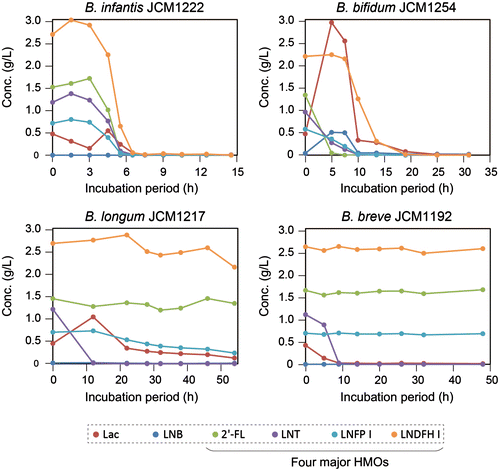
As expected, B. infantis imported a wide variety of intact HMOs (Fig. ). All oligosaccharides disappeared when the organism entered the logarithmic phase, which was followed by a temporal increase in a small amount of monosaccharides (Fuc, Gal, and Glc). The transient increase in monosaccharides might be a counteraction against the osmotic pressure caused by the rapid uptake of large amounts of HMOs. The HMO consumption behavior of B. infantis is consistent with the metabolic ability deduced by the genomic analysis.Citation55) B. infantis encodes 20 genes of family 1 solute-binding proteins of the ABC transporter, seven of which are located in the HMO cluster-1. Based on a glycan array analysis, Garrido et al. showed that some of the isolated solute-binding proteins indeed recognize the glycan structures found in HMOs, although further biochemical studies are needed to clarify which transporter is exactly involved in the uptake of each of the HMOs.Citation70) B. infantis is equipped with all types of glycosidases required to hydrolyze the linkages of HMOs in its cytoplasm, and thus consumed all HMOs from the medium.Citation55,56,62) B. bifidum, another avid HMO consumer, was found to degrade HMOs in a different manner. As 1,2-α-L-fucosidase, 1,3-1,4-α-L-fucosidase, and LNBase of B. bifidum are secretory enzymes,Citation9,62) the organism degraded HMOs into mono- and disaccharides extracellularly. Unexpectedly, this degradation occurred before the species entered the exponential growth phase (Fig. ). The degraded mono- (Fuc, Gal, and Glc) and disaccharides (LNB and Lac) were abundantly present in the culture medium of B. bifidum between the early and mid-logarithmic phases.Citation24) Even after degrading all kinds of HMOs and entering the stationary phase, monosaccharides remained unconsumed. Similar to B. longum, B. bifidum also has a preference for LNB over Lac. Nonetheless, the LNB concentration (1.3 mM) at the lag phase (before entering logarithmic phase) reached to half of the sum of the initial concentrations of LNT and LNFP I. The Lac concentration became equal to the sum of LNT and LNFP I. These results suggest that these metabolites are shared among gut microbes, especially among bifidobacteria.Citation24) Importantly, the GNB/LNB transporter, GNB/LNB phosphorylase (GLNBP), and LNB assimilation ability are limited to infant-gut-associated bifidobacteria.Citation71,72) B. infantis appears to be “selfish” in utilizing HMOs, while B. bifidum is “altruistic.” It is interesting to note that Tannock et al. found that the population and diversity of bifidobacteria increase significantly when B. bifidum occupies > 10% of the total bifidobacterial counts in infant feces, as compared with the case that B. bifidum population counts for less than 10%.Citation37) They also mentioned that this phenomenon only occurs in breast-fed babies, strongly suggesting the symbiotic sharing of HMOs among bifidobacteria.
B. infantis and B. bifidum were able to consume type-2 HMOs efficiently.Citation24,73) The hydrolysis of type-2 oligosaccharides occurs, respectively, inside and outside of B. infantis and B. bifidum cells. Our group has already identified β-galactosidase that specifically acts on lacto-N-neotetraose (Galβ1-4GlcNAcβ1-3Galβ1-4Glc)Citation56) and β-HexNAc-ase that is highly specific for lacto-N-triose II (GlcNAcβ1-3Galβ1-4Glc).Citation73)
VI. Concluding remarks
The gut microbial population of breast-fed infants constitutes a tripartite relationship between the mother’s milk, the infant, and bacteria. In general, bifidobacteria constitute a larger part of the microbiota of breast-fed newborns than that of bottle-fed babies.Citation13–15) Albrecht et al. examined the profiles of the oligosaccharides present in the milk of breast-feeding mothers and the feces of the corresponding infants and found that the HMO degradation in the infant’s digestive tract is highly variable.Citation74) In one case, an oligosaccharide lacking the LNB moiety of fucosyllacto-N-hexaose II was identified in the infant feces (and was not present in the mother’s sample), which strongly suggests the action of LNBases in the intestine of the infant. In the same sample, a reduction in fucosylated oligosaccharides and LNT was observed. Although the oligosaccharide profile in milk did not change significantly during the first six months after giving a birth, the profiles of infant feces showed drastic changes. This indicates that the changes in oligosaccharide degradation in the infant digestive tract are linked with a maturation process of gut microbiota.Citation74) De Leoz et al. have recently confirmed that infants with bifidobacteria-rich gut microbiota exhibit a more extensive HMO degradation in their feces than infants with microbiota with less bifidobacteria, although this study only compared the feces of two babies.Citation75) In combination with the above-mentioned findings, it is therefore highly likely that HMOs serve to stimulate the growth of bifidobacteria to dominate the infant gut microbial population.
In retrospect, the 1954 report by Gauhe et al. describing oligosaccharides consisted of Fuc, Gal, Glc, and GlcNAc that act as bifidus factor was fairly accurate.Citation22) Although their results were dismissed based on the use of a variant (perhaps, mutant) strain of bifidobacteria, they already mentioned that bifidobacterial strains with the same phenotype (i.e., requiring HMOs for rapid growth, but not cow milk) were frequently isolated from the stool of breast-fed babies. Unfortunately, these pioneering findings could not be validated at the time because of a lack of genetic and analytical tools. In addition, it was difficult to conceive that the specific assimilation of HMOs by bifidobacteria involves and relies on the uptake of di- or longer saccharides (LNB, LNT, LNFP I, etc.).
HMOs have no apparent nutritional value for infants, even though they are the third most abundant component in human milk. Nevertheless, human females have evolved to produce a large amount of HMOs (10–20 g/L) in the mammary glands at a great expense of energy.Citation29) Apart from the structural diversity among mammals, milk oligosaccharides also appeared in monotreme species (platypus and echidna).Citation76) Urashima et al. have claimed that the original function of milk oligosaccharides is the prevention of microbial infection (HMOs also work in that way) and that the roles of milk oligosaccharides have diversified later during evolution.Citation76) In a rare case of cross-kingdom symbiosis, humans and bifidobacteria might have succeeded in mutually exploiting HMOs as beneficial compounds, as deduced by the fact that the presence of type-1 chain-specific enzymes is limited to infant-gut-associated bifidobacteria and that only human milk is rich in type-1 chain oligosaccharides.
Funding
The research described herein was supported in part by a Grant-in-Aid for Scientific Research (B) 15H04481 from the Ministry of Education, Culture, Sports, Science and Technology, Japan; a Grant-in-Aid from the Asahi Glass Foundation, the Urakami Foundation, and Nagase Science Technology Foundation; a Grant-in-Aid from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, Japan; and a Grant-in-Aid from the Institution for Fermentation, Osaka, Japan.
Disclosure statement
No potential conflict of interest was reported by the author.
Acknowledgments
Some parts of this manuscript have been published in the feature article (in Japanese) of the Japanese Society of Applied Glycoscience and also will appear in the short review (in Japanese) of Milk Science. Most of the research described in this manuscript was carried out in collaboration with Drs. Motomitsu Kitaoka (National Food Research Institute, National Agriculture and Food Research Organization), Shinya Fushinobu (The University of Tokyo), and Hisashi Ashida (Kinki University). Our work is deeply indebted to the pioneering research on milk oligosaccharides of Dr. Tadasu Urashima of the Obihiro University of Agriculture and Veterinary Medicine. Special thanks go out to Drs. Junko Hirose, Sadaki Asakuma, Jun Wada, Masashi Kiyohara (departed colleague), Haruko Sakurama, Erina Yoshida, Aina Gotoh, Alexander H. Viborg, and Birte Svensson. Finally, I express my sincere gratitude to Drs. Hidehiko Kumagai and Kenji Yamamoto of Ishikawa Prefectural University (both Professor Emeritus at Kyoto University) for their continuous encouragement, and to Drs. Hideyuki Suzuki (Kyoto Institute of Technology), Hisanori Tamaki (Kagoshima University), and Hiromichi Minami (Ishikawa Prefectural University) for their helpful advice.
Notes
† This review was written in response to the author’s receipt of the Japan Society for Bioscience, Biotechnology, and Agrochemistry Award for the Encouragement of Young Scientists in 2011.
References
- Schloissnig S, Arumugam M, Sunagawa S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50.
- Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286.10.1038/nature08530
- Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450.10.1038/nature12721
- Tobisawa Y, Imai Y, Fukuda M, et al. Sulfation of colonic mucins by N-acetylglucosamine 6-O-sulfotransferase-2 and its protective function in experimental colitis in mice. J. Biol. Chem. 2010;285:6750–6760.10.1074/jbc.M109.067082
- Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463.10.1016/j.cell.2013.11.024
- Nakayama J, Watanabe K, Jiang J, et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015;5:8397.10.1038/srep08397
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563.
- Larsbrink J, Rogers TE, Hemsworth GR, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502.10.1038/nature12907
- Katayama T, Sakuma A, Kimura T, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 2004;186:4885–4893.10.1128/JB.186.15.4885-4893.2004
- Fujita K, Oura F, Nagamine N, et al. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 2005;280:37415–37422.10.1074/jbc.M506874200
- Katayama T, Fujita K, Yamamoto K. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J. Biosci. Bioeng. 2005;99:457–465.10.1263/jbb.99.457
- Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13.10.1186/s40168-015-0071-z
- Mitsuoka T. Intestinal flora and host. Asian Med. J. 1988;31:400–409.
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227.
- Sakata S, Tonooka T, Ishizeki S, et al. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 2005;243:417–423.10.1016/j.femsle.2005.01.002
- Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521.10.1542/peds.2005-2824
- Enomoto T, Sowa M, Nishimori K, et al. Effects of bifidobacterial supplementation to pregnant women and infants in the prevention of allergy development in infants and on fecal microbiota. Allergol. Int. 2014;63:575–585.10.2332/allergolint.13-OA-0683
- Ishizuka S, Iwama A, Dinoto A, et al. Synbiotic promotion of epithelial proliferation by orally ingested encapsulated Bifidobacterium breve and raffinose in the small intestine of rats. Mol. Nutr. Food Res. 2009;53(S1):S62–S67.10.1002/mnfr.v53.5s
- Groeger D, O’Mahony L, Murphy EF, et al. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes. 2013;4:325–339.10.4161/gmic.25487
- György P, Norris RF, Rose CS. Bifidus factor. I. a variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 1954;48:193–201.10.1016/0003-9861(54)90323-9
- György P, Kuhn R, Rose CS, et al. Bifidus factor. II. its occurrence in milk from different species and in other natural products. Arch. Biochem. Biophys. 1954;48:202–208.10.1016/0003-9861(54)90324-0
- Gauhe A, György P, Hoover JR, et al. Bifidus factor. IV. preparations obtained from human milk. Arch. Biochem. Biophys. 1954;48:214–224.10.1016/0003-9861(54)90326-4
- Glick MC, Sall T, Zilliken F, et al. Morphological changes of Lactobacillus bifidus var. pennsylvanicus produced by a cell-wall precursor. Biochim. Biophys. Acta. 1960;37:361–363.10.1016/0006-3002(60)90251-1
- Asakuma S, Hatakeyama E, Urashima T, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011;286:34583–34592.10.1074/jbc.M111.248138
- Ruiz-Palacios GM, Cervantes LE, Ramos P, et al. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003;278:14112–14120.10.1074/jbc.M207744200
- Crane JK, Azar SS, Stam A, et al. Oligosaccharides from human milk block binding and activity of the Escherichia coli heat-stable enterotoxin (STa) in T84 intestinal cells. J. Nutr. 1994;124:2358–2364.
- Hong P, Ninonuevo MR, Lee B, et al. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br. J. Nutr. 2009;101:482–486.
- Jantscher-Krenn E, Zherebtsov M, Nissan C, et al. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut. 2012;61:1417–1425.10.1136/gutjnl-2011-301404
- Urashima T, Kitaoka M, Terabayashi T, et al. Milk oligosaccharides. New York, NY: Nova Science Publishers; 2011. p. 1–93.
- Ninonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 2006;54:7471–7480.10.1021/jf0615810
- Kobata A. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:731–747.10.2183/pjab.86.731
- Kunz C, Rudloff S, Baier W, et al. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722.10.1146/annurev.nutr.20.1.699
- Brand-Miller JC, McVeagh P, McNeil Y, et al. Digestion of human milk oligosaccharides by healthy infants evaluated by the lactulose hydrogen breath test. J. Pediatr. 1998;133:95–98.10.1016/S0022-3476(98)70185-4
- Dotz V, Rudloff S, Meyer C, et al. Metabolic fate of neutral human milk oligosaccharides in exclusively breast-fed infants. Mol. Nutr. Food Res. 2015;59:355–364.10.1002/mnfr.201400160
- Ruhaak LR, Stroble C, Underwood MA, et al. Detection of milk oligosaccharides in plasma of infants. Anal. Bioanal. Chem. 2014;406:5775–5784.10.1007/s00216-014-8025-z
- He Y, Liu S, Kling DE, et al, The human milk oligosaccharide 2′-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation, Gut 2016;65:33–46.
- Tannock GW, Lawley B, Munro K, et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk, Appl. Environ. Microbiol. 2013;79:3040–3048.10.1128/AEM.03910-12
- Derensy-Dron D, Krzewinski F, Brassart C, et al. Beta-1,3-galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol. Appl. Biochem. 1999;29(Pt 1):3–10.
- Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 2005;71:3158–3162.10.1128/AEM.71.6.3158-3162.2005
- Wada J, Ando T, Kiyohara M, et al. Bifidobacterium bifidum Lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 2008;74:3996–4004.10.1128/AEM.00149-08
- Sano M, Hayakawa K, Kato I. An enzyme releasing lacto-N-biose from oligosaccharides. Proc. Natl. Acad. Sci. USA. 1992;89:8512–8516.10.1073/pnas.89.18.8512
- Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644.10.1016/S0959-440X(97)80072-3
- Mark BL, Mahuran DJ, Cherney MM, et al. Crystal structure of human β-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay–Sachs disease. J. Mol. Biol. 2003;327:1093–1109.10.1016/S0022-2836(03)00216-X
- Mark BL, Vocadlo DJ, Knapp S, et al. Crystallographic evidence for substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 2001;276:10330–10337.10.1074/jbc.M011067200
- Lemieux MJ, Mark BL, Cherney MM, et al. Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 2006;359:913–929.10.1016/j.jmb.2006.04.004
- Ito T, Katayama T, Hattie M, et al. Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum. J. Biol. Chem. 2013;288:11795–11806.10.1074/jbc.M112.420109
- Hattie M, Ito T, Debowski AW, et al. Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis. Chem. Commun. (Camb). 2015;51:15008–15011.10.1039/C5CC05494J
- Fukuda S, Toh H, Taylor TD, et al. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449–454.10.4161/gmic.21214
- Honda Y, Nishimoto M, Katayama T, et al, Characterization of the cytosolic β-N-acetylglucosaminidase from bifidobacterium longum subsp. longum. J. Appl. Glycosci, 60, 141–146 (2013).10.5458/jag.jag.JAG-2013_001
- Sakurama H, Kiyohara M, Wada J, et al. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J. Biol. Chem. 2013;288:25194–25206.10.1074/jbc.M113.484733
- Gotoh A, Katoh T, Sugiyama Y, et al. Novel substrate specificities of two lacto-N-biosidases towards beta-linked galacto-N-biose-containing oligosaccharides of globo H, Gb5, and GA1. Carbohydr. Res. 2015;408:18–24.10.1016/j.carres.2015.03.005
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410.10.1016/S0022-2836(05)80360-2
- Suzuki R, Wada J, Katayama T, et al. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 2008;283:13165–13173.10.1074/jbc.M709777200
- Tam R, Saier MH Jr. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 1993;57:320–346.
- Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:18964–18969.10.1073/pnas.0809584105
- Yoshida E, Sakurama H, Kiyohara M, et al. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–368.10.1093/glycob/cwr116
- Viborg AH, Fredslund F, Katayama T, et al, A beta1-6/beta1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium, Mol. Microbiol 2014;94:1024–1040.
- Viborg AH, Katayama T, Abou Hachem M, et al. Distinct substrate specificities of three glycoside hydrolase family 42 beta-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology. 2014;24:208–216.10.1093/glycob/cwt104
- Nagae M, Tsuchiya A, Katayama T, et al. Structural basis of the catalytic reaction mechanism of novel 1,2-α-L-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 2007;282:18497–18509.10.1074/jbc.M702246200
- Wada J, Honda Y, Nagae M, et al. 1,2-alpha-l-Fucosynthase: a glycosynthase derived from an inverting alpha-glycosidase with an unusual reaction mechanism. FEBS Lett. 2008;582:3739–3743.10.1016/j.febslet.2008.09.054
- Ashida H, Miyake A, Kiyohara M, et al. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017.10.1093/glycob/cwp082
- Sakurama H, Fushinobu S, Hidaka M, et al. 1,3-1,4-α-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J. Biol. Chem. 2012;287:16709–16719.10.1074/jbc.M111.333781
- Sakurama H, Tsutsumi E, Ashida H, et al. Differences in the substrate specificities and active-site structures of two α-L-fucosidases (glycoside hydrolase family 29) from bacteroides thetaiotaomicron. Biosci. Biotechnol. Biochem. 2012;76:1022–1024.10.1271/bbb.111004
- Rodriguez-Diaz J, Rubio-del-Campo A, Yebra MJ. Lactobacillus casei ferments the N-Acetylglucosamine moiety of fucosyl-alpha-1,3-N-acetylglucosamine and excretes L-fucose. Appl. Environ. Microbiol. 2012;78:4613–4619.10.1128/AEM.00474-12
- Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv. Nutr. 2012;3:422S–429S.10.3945/an.111.001420
- Odamaki T, Horigome A, Sugahara H, et al. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int. J. Genomics. 2015:567809.
- Ward RE, Niñonuevo M, Mills DA, et al. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol. Nutr. Food Res. 2007;51:1398–1405.10.1002/(ISSN)1613-4133
- LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007;55:8914–8919.10.1021/jf0710480
- Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010;58:5334–5340.10.1021/jf9044205
- Garrido D, Kim JH, German JB, et al. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One. 2011;6:e17315.10.1371/journal.pone.0017315
- Kiyohara M, Tachizawa A, Nishimoto M, et al. Prebiotic effect of lacto-N-biose I on bifidobacterial growth. Biosci. Biotechnol. Biochem. 2009;73:1175–1179.10.1271/bbb.80697
- Xiao JZ, Takahashi S, Nishimoto M, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 2010;76:54–59.10.1128/AEM.01683-09
- Miwa M, Horimoto T, Kiyohara M, et al. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20:1402–1409.10.1093/glycob/cwq101
- Albrecht S, Schols HA, van den Heuvel EG, et al. Occurrence of oligosaccharides in feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carbohydr. Res. 2011;346:2540–2550.10.1016/j.carres.2011.08.009
- De Leoz ML, Kalanetra KM, Bokulich NA, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J. Proteome Res. 2015;14:491–502.10.1021/pr500759e
- Urashima T, Inamori H, Fukuda K, et al. 4-O-Acetyl-sialic acid (Neu4,5Ac2) in acidic milk oligosaccharides of the platypus (Ornithorhynchus anatinus) and its evolutionary significance. Glycobiology. 2015;25:683–697.10.1093/glycob/cwv010
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948.
