Abstract
SRF-MADS proteins are transcription factors conserved among eukaryotes that regulate a variety of cellular functions; however, their physiological roles are still not well understood in filamentous fungi. Effects of a mutation in mcmA gene that encodes the sole SRF-MADS protein in the fungus Aspergillus nidulans were examined by RNA sequencing. Sequencing data revealed that expression levels of cellulase genes were significantly decreased by the mutation as reported previously. However, expression levels of various hemicellulolytic enzyme genes, several extracellular protease genes, the nosA and rosA genes involved in sexual development, and AN4394 encoding an ortholog of EcdR involved in Aspergillus oryzae conidiation, were also significantly decreased by the mutation. As expected from the RNA sequencing data, the mcmA mutant had reduced protease production, cleistothecial development, and conidiation. This is the first report describing the involvement of SRF-MADS proteins in protease production in fungi, and asexual and sexual development in Aspergillus.
Graphical abstract
McmA regulated cellulase/protease production and asexual/sexual development. The figure shows impaired development of fruiting bodies (cleistothecia) in the mcmA mutant.
MADS box proteins are transcription factors conserved among eukaryotes that are involved in the regulation of a wide variety of cellular functions. They function in concert with various cofactors, such that the target genes vary depending on the cofactor employed.Citation1) There are two types of MADS proteins, SRF-type and MEF2-type, which are classified based on the amino acid sequence of the conserved MADS box. Saccharomyces cerevisiae expresses two SRF-type proteins, MCM1 and ARG80, and two MEF2-type proteins, RLM1 and SMP1. These proteins regulate mating-type specific genes (MCM1), as well as genes regulating cell cycle (MCM1), arginine metabolism (MCM1 and ARG80), osmotic stress response (SMP1), and cell wall integrity (RLM1).Citation2–7)
The homologs of these proteins are currently not well-characterized in filamentous ascomycetes. In the case of SRF-type MADS proteins, MCM1 regulates fruiting body development in Sordaria macrospora, and MoMcm1 regulates male fertility, microconidium production, and virulence in Magnaporthe oryzae.Citation8,9) In A. nidulans, McmA regulates expression of cellulase genes.Citation10) MADS1 in Fusarium verticillioides is involved in the regulation of secondary metabolism,Citation11) and MadsA controls the phase transition from yeast to mycelium in Penicillium marneffei.Citation12) Among MEF-type MADS proteins, RlmA regulates α-1,3-glucan synthesis in A. niger and A. nidulans.Citation13,14) Mig1 is required for infectious growth in Magnaporthe grisea.Citation15)
These reports suggest that MADS box proteins in filamentous ascomycetes regulate a variety of cellular functions as in S. cerevisiae, however, studies to identify the multiple roles of a single MADS box protein in a single fungal species have not been performed. In this study, using RNA sequencing, we demonstrate the involvement of A. nidulans McmA in the regulation of cellulase, hemicellulase, and protease production, as well as asexual and sexual development.
Materials and methods
A. nidulans strains and cultural conditions
A. nidulans D6B (biA1 pyrG89; wA3; argB2::argB::eglAp-taaG2; pyroA4),Citation16) the mcmAI70A mutant (MCMI70A) (biA1 pyrG89; wA3; ΔmcmA::mcmAI70A::pyr4 argB2::argB::eglAp-taaG2; pyroA4), and MCMI70AC (biA1 pyrG89; wA3; ΔmcmA::mcmAI70A::pyr4 argB2::argB::eglAp-taaG2; pyroA4::pyroA::mcmA+) were used in this study.Citation10,16) The strains were grown at 37 °C in standard minimal medium (SMM) with appropriate supplements unless otherwise noted.Citation17)
RNA sequencing
A. nidulans D6B and MCMI70A strains were pre-cultured for 24 h at 37 °C in SMM with 2% glucose as the carbon source. The mycelia were collected and washed with SMM-lacking carbon sources. The mycelia (0.5 g) were transferred to 40 ml of fresh SMM containing 0.1% cellobiose as the carbon source, and cultured at 37 °C. The mycelia were harvested at 3 h after inoculation, frozen in liquid nitrogen, and broken with an SK-mill (SK-100, Tokken, Japan). RNA was extracted from the mycelia using the RNeasy Mini Kit (Qiagen) according to the supplier’s instructions.
RNA sequencing was performed by Genaris, Inc. (Kanagawa, Japan) according to Illumina protocols, and the reads were mapped against the A. nidulans genome sequence at the Broad Institute (http://www.broadinstitute.org) with TopHat.Citation18) Transcripts were quantified as FPKM (Fragments Per Kilobase of transcript per Million mapped reads) using Cufflinks.Citation19)
Protease assay
The plate assay for protease production was performed by observing a zone of clearance (halo) around a colony formed by spot inoculation of 106 conidiospores from each strain on SMM agar with 1% casein as the sole carbon source. Production of protease in submerged cultures was determined as follows. A. nidulans strains were pre-cultured for 20 h in SMM with 1% glucose as the sole carbon source, and then washed with the same medium without a carbon source. The mycelia of approximately 0.5 g in wet weight were then transferred to 40 ml of SMM with and without a carbon source and further cultivated for 16 h. Carbon sources used included casein, glucose, or cellobiose, at a final concentration of 1%.
The protease assay of the culture supernatants at 16 h was carried out at 37 °C using azocasein as the substrate using a method described previously.Citation20) The reaction mixture contained 0.25% azocasein in 50 mM phosphate buffer, pH 7.2. Protease activity was measured as the increase in absorbance at 440 nm per min and per gram dry mycelium.
Asexual and sexual development
The development of asexual spores (conida) in A. nidulans strains was examined by counting the number of conidia under a microscope. A. nidulans strains were grown on SMM agar plates for 5 days, and conidia were collected as a suspension in a solution containing 0.1% Tween 80 and 0.9% NaCl. The number of conidia per square centimeter of the plate culture was determined.
In the examination of sexual development, A. nidulans strains were grown at 37 °C for 2 days on SMM agar plates. The plates were then sealed to cause hypoxic conditions and further incubated at 37 °C for 14 days in the dark to induce the formation of cleistothecia (fruiting bodies). Microscopic observation was carried out using the SteREO Lumar.V12 (Carl Zeiss, Jena, Germany) stereomicroscope.
RNA preparation and Northern blotting analysis
To obtain RNA at the asexual development stage, A. nidulans mycelia grown in SMM submerged culture at 37 °C for 22 h were collected, spread onto SMM agar plates, and further cultivated at 37 °C. Growth conditions to detect the protease gene (prtA) transcripts were identical to those used for the protease assay.
RNA was extracted from A. nidulans by TRIzol® (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s protocol. Total RNA (12 μg) was used in Northern blot analysis, which was performed as previously described.Citation21) Probes were amplified using the PCR DIG Probe Synthesis Kit (Roche, Mannheim, Germany) with A. nidulans cDNA as the template. Primers used for amplification are listed in Table S1. Hybridization signals were detected using CSPD (Roche Applied Science, Penzberg, Germany).
Results
Genome-wide screening for possible McmA targets by RNA sequencing
In a previous study, we demonstrated that a mutation in the mcmA gene encoding the SRF-MADS protein McmA causes impaired cellulase production on carboxymethyl cellulose (CMC) and decreased expression of the cellulase genes eglA, eglB, and cbhA.Citation10) To better understand McmA-mediated gene regulation, the effect of the mcmA mutation on genome-wide gene expression was analyzed by RNA sequencing. RNA was extracted from the mycelia of D6B (mcmA+) and MCMI70A (mcmAI70A) after 3 h of cellobiose induction that aimed to maximize the expression of genes encoding cellulolytic enzymes. Total numbers of the mapped reads were 101,092,198 for D6B and 81,263,650 for MCMI70A, and average expression levels were 69.61 in D6B and 70.09 FPKM in MCMI70A (Table S2). The 7149 genes that had FPKM values of over 5 in D6B were selected for further analysis.
A more than fourfold decrease of FPKM in MCMI70A was observed in 208 genes compared to those in D6B (Table S2). Among them, 104 genes that had FPKM values above 50 in D6B are listed in Table . These include the eglA (AN1285), eglB (AN3418), and cbhA (AN5176) genes, which have been shown to be regulated by McmA, and six additional genes that are possibly involved in cellulose degradation, including three putative lytic polysaccharide monooxygenase genes (AN3860, AN6428, AN2388), a putative cellobiohydrolase gene cbhD (AN1273), and two putative β-glucosidase genes, AN10124 and bglI (AN2227). Except for AN6428 and AN2388, all were highly expressed in D6B, with FPKM values over 500, suggesting that McmA plays an essential role in cellulose degradation. Since there was a possibility that some highly expressed cellulolytic enzyme genes were missing due to low dependency on McmA, we searched for possible cellulolytic enzyme genes with high FPKM values (Table S2). No putative cellulolytic enzyme genes with an FPKM value over 500 were present.
Table 1. Possible targets of McmA. Genes with FPKM values of over 50 in D6B and D6B/I70A FPKM ratio of over 4 are shown. The data on the genes with FPKM below 50 is available from the supplementary Table 1. Genes encoding carbohydrate-active enzymes are shadowed. Descriptions of the genes were obtained from AspGD web site.
In the course of the study on regulation of cellulolytic enzyme genes in N. crassa as well as in A. nidulans, Coradetti et al. have reported that the transcription factor ClrB in A. nidulans is essential for the cellulose-induced expression of cellulolytic enzyme genes as well as a number of hemicellulolytic enzyme genes, and that a total of 141 genes are under its control.Citation22) Comparison of the McmA-regulated and ClrB-regulated genes revealed that 17 genes were cooperatively regulated by McmA and ClrB, including the 7 highly expressed cellulolytic enzyme genes (Table ). However, no endoxylanase and β-xylosidase genes were found to be coregulated. Although a single endomannanase gene manB was shown to be regulated by both McmA and ClrB, no β-mannosidase genes were regulated by McmA (Table S2). In addition, although the highly expressed endomannanase genes reported are manC (AN6427) and man134A (AN2710),Citation23) they were not regulated by McmA (Table S2). These results imply that cooperative regulation by McmA and ClrB is essential for cellulose degradation and is partially involved in hemicellulose degradation in A. nidulans.
Table 2. Genes possibly regulated by both McmA and ClrB. Descriptions of the genes were obtained from AspGD web site.
Besides the polysaccharide-degrading enzyme genes, the RNA sequencing analysis revealed a possible involvement of McmA in protease production and asexual/sexual development, based on the decrease in expression levels of prtA, nosA, rosA, and AN4394 in the mcmA mutant (Table ). The prtA gene encodes the protease PrtA in A. nidulans,Citation24) nosA and rosA encode transcription factors involved in regulation of sexual development,Citation25,26) and EcdR, the A. oryzae ortholog of AN4394, regulates early asexual development.Citation27)
Regulation of protease production by McmA
A. nidulans produces an extracellular proteinase, PrtA, as a predominant proteolytic enzyme.Citation24) RNA sequencing analysis revealed expression levels of prtA, as well as other protease genes, namely AN7159 (putative tripeptidyl peptidase), AN2555 (putative carboxypeptidase), and pepJ (deuterolysin-type metalloproteinase),Citation28) were significantly decreased in the mcmA mutant (Table , Table S2). As shown in Fig. (A), while D6B produced a halo around the colony on agar with casein as the sole carbon source, the mcmA mutant MCMI70A failed produce a halo, indicating that protease production was impaired by the mutation. Reinsertion of the wild type mcmA gene into the mutant restored the protease production (MCMI70AC).
Fig. 1. Protease production in the parental strain D6B and the mcmA mutant.
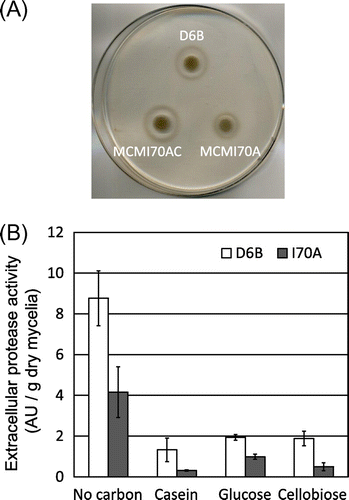
Since expression of prtA is reported to be induced under starvation conditions,Citation29) protease production was examined using various carbon sources as well as carbon-depleted conditions (Fig. (B)). Carbon depletion led to a significantly higher protease production in the D6B strain and the mcmA mutation caused reduction of production by approximately 50%. Protease production in D6B using casein, glucose, and cellobiose was low compared to the carbon-depleted conditions, and the mcmA mutation caused further decrease in production.
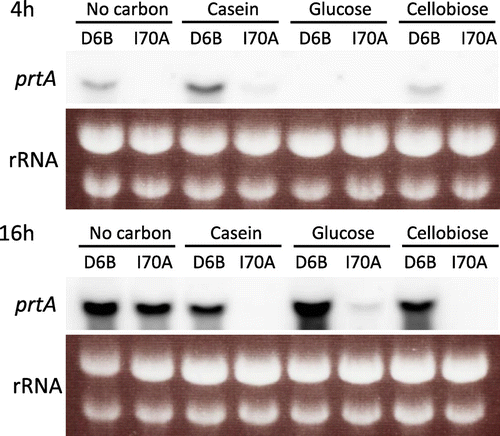
Effect of the mcmA mutation on the prtA expression was examined by Northern blot analysis (Fig. ). At 4 h after the transfer of the pre-grown mycelia to fresh media, detectable amounts of the prtA mRNA accumulated in D6B, except when glucose was used, which is likely due to carbon catabolite repression. The mcmA mutation caused a decrease in prtA transcription under all the conditions examined. At 16 h after the transfer, the D6B strain accumulated prtA mRNA under all the conditions, and the mcmA mutation caused a significant decrease in expression in casein, glucose, and cellobiose. Strikingly, significant amounts of prtA mRNA accumulated in the mcmA mutant under carbon-depleted conditions, although the mRNA level was lower than that in D6B. These observations imply that mcmA is involved in regulation of protease genes, however, it may not be necessary in induction of prtA expression under carbon-starved conditions.
Fig. 2. Expression of the prtA gene with various carbon sources and effect of the mcmA mutation on its expression.
Despite the accumulation of prtA mRNA in D6B under all the conditions at 16 h, protease production on casein, glucose, and cellobiose was significantly lower than that under carbon-starved conditions (Fig. (B)). Such discrepancy might be attributable to differences in the time-course profiles of prtA mRNA accumulation; in other words, the final amount of PrtA in the culture broth is affected by the time when the transcription is triggered during the course of cultivation.
Involvement of McmA in regulation of sexual development
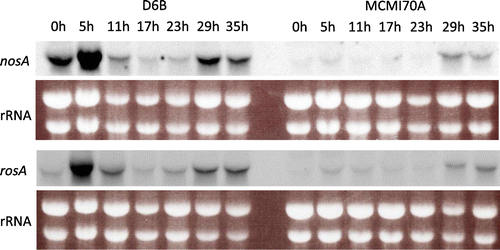
Expression of the nosA and rosA genes, which encode transcription factors involved in sexual development,Citation25,26) was significantly decreased by the mcmA mutation according to the RNA sequencing analysis (Table ). As nosA and rosA expression is upregulated during late asexual development, Northern blot analysis was performed with the mycelia grown on agar plates as described previously.Citation25,26) As shown in Fig. , the nosA and rosA transcripts in D6B transiently increased at the initial 5 h stage, then decreased for 24 h, and again increased at 29 h and 35 h. In the mcmA mutant, significantly decreased nosA and rosA expression was observed throughout the cultivation, confirming that McmA regulates expression of these two genes.
Fig. 3. Time course expression of nosA and rosA genes and effect of the mcmA mutation on their expression.
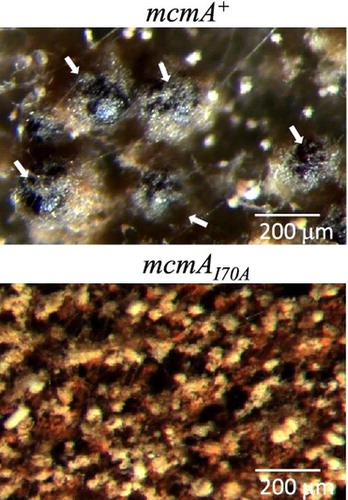
As a consequence of sexual development, A. nidulans forms spherical fruiting bodies called cleistothecia, which contain ascospores. To assess the effect of the mcmA mutation on sexual development, cleistothecial formation by the mutant strain was examined. While the strain D6B formed cleistothecia with a blackball-like appearance, no cleistothecial formation was observed in the mcmA mutant (Fig. ).
Involvement of McmA in regulation of asexual development
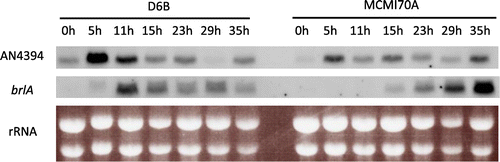
Expression of AN4394, which is an ortholog of ecdR in A. oryzae,Citation27) was found to be significantly decreased in the mcmA mutant based on RNA sequencing analysis (Table ). The ecdR gene encodes the basic helix–loop–helix transcription factor involved in the early stage of conidiophore development in A. oryzae, which suggests that McmA regulates asexual development via regulation of AN4394. Furthermore, decrease in the expression of brlA, the C2H2 zinc finger transcription factor involved in conidiophore development,Citation30) was also observed in the mcmA mutation (Table S2). Examination of the conidiospore-forming activity of D6B and the mcmA mutant revealed a ~50% decrease in conidiospore formation in the mutant, that is, while D6B formed (1.05 ± 0.03) × 108 spores/cm2, the mutant formed (0.53 ± 0.06) × 108 spores/cm2.
Expression levels of AN4394 and brlA under plate culture conditions were determined by Northern blot analysis. In the D6B strain, expression of AN4394 was transiently enhanced at an initial stage, with a peak at 5 h after the transfer of the mycelia on to the agar plate, and expression decreased afterward (Fig.). The brlA expression occurred just after the peak expression of AN4394 with a peak at 11 h and then decreased afterward, suggesting that AN4394 enhances expression of brlA. In the mcmA mutant, the transiently enhanced expression of AN4394 was weak, and expression of brlA was significantly delayed, potentially leading to the delayed formation of conidiospores.
Discussion
We have reported in a previous study that the SRF-MADS box protein McmA in A. nidulans regulates the endoglucanse genes eglA and eglB, as well as the putative cellobiohydrolase gene cbhA.Citation10) Our study revealed that there were seven highly expressed cellulolytic enzyme genes when cellobiose was the carbon source, and that all these genes were regulated by McmA. Our findings indicated the essential role of McmA in cellulose degradation. Furthermore, we found McmA also regulated protease production and asexual/sexual development.
SRF-MADS proteins function in concert with cofactors, which are mostly transcription factors.Citation1) In the case of regulation of cellulolytic enzyme genes, the most likely McmA partner is ClrB, based on studies of the eglA promoter. This promoter has a single cis-element, designated CeRE,Citation16) that is required for inductive expression, and thus ClrB may bind to this element to regulate induction. Since McmA does bind to CeRE,Citation10) it is highly possible that the gene is regulated through interaction between McmA and ClrB. It should be noted that only 17 genes in Table showed McmA dependency among the 141 ClrB-regulated genes.Citation22) Regulatory systems mediated by ClrB may be similar to those regulated by Ste12p in S. cerevisiae, in which, while Ste12p alone regulates pheromone-regulated genes, it also coregulates a-specific genes with Mcm1p.Citation31) Strikingly, AN2184, an ortholog of Neurospora crassa cdt1 that encodes a cellobiose transporter,Citation32) was not regulated by McmA, although its expression level was extremely high, with FPKM of over 3000 in the D6B strain (Table S2). This suggests that the role of McmA is limited to cellulose hydrolysis, while ClrB regulates the whole system of cellulose utilization.
As is the case for regulation of cellulolytic genes, an McmA cofactor is likely to exist for the regulation of protease production and asexual/sexual development. Currently identified transcriptional activators, AreA, XprG, and PacC, might function as cofactors in the regulation of the extracellular protease gene prtA.Citation29,33,34) In A. niger, the induction of extracellular protease is regulated by the transcriptional activator PrtT.Citation35) However, A. nidulans lacks an ortholog of prtT. Consistent with the lack of prtT, extracellular proteases are not induced by exogenous proteins, instead they are produced under carbon-, nitrogen- or sulfur-limiting conditions and at neutral to alkaline pH.Citation34,36) AreA and XprG are responsible for prtA expression under nitrogen starvation, XprG is responsible under carbon starvation, and PacC is responsible at higher pH conditions.Citation29,34,37) McmA does not seem to regulate these transcription factor genes since the expression of areA, xprG, and pacC was not significantly affected by the mcmAI70A mutation (Table S2). It is possible that one of these factors may regulate prtA expression in concert with McmA.
Among the many transcription factors involved in regulation of sexual development,Citation38) nosA and rosA gene expression was greatly decreased by the mcmA mutation. NosA is a Zn(II)2Cys6 transcription factor that promotes cleistothecial formation.Citation25) Although no transcription factor has been identified that activates nosA expression, NsdD might be involved in cooperative regulation with McmA, as it has been suggested that NosA acts in the same pathway downstream of NsdD or in a parallel pathway.Citation25) Another possible McmA partner is SteA, which is also required for cleistothecial development.Citation39) The orthologs of McmA and SteA in S. cerevisiae, Mcm1p and Ste12p, physically interact and regulate mating type-specific genes.Citation40) Interaction of the orthologs in Sordaria macrospora, MCM1 and STE12, was also reported. In this organism, MCM1 but not STE12 is required for fruiting body development, while STE12 deletion affects the ascus and ascospore development, implying functional divergence within the fungal STE12 transcription factor family.Citation8,41) The rosA gene is a paralog of nosA, however, it encodes a repressor of sexual development.Citation26) In our experimental conditions, their expression profiles, along with growth, were nearly identical and both displayed McmA dependency for their expression (Fig. ). This indicates the presence of transcripts for an activator and a repressor at the same time, suggesting that external stimuli might regulate translation or activity of these proteins.
The mcmA mutation caused decreased expression of AN4394 (Table ), an ortholog of ecdR in A. oryzae,Citation27) as well as decreased conidiospore formation. The deletion of ecdR in A. oryzae causes a decrease in expression of brlA, which is essential to conidiation in A. nidulans.Citation30) Consistent with this, brlA expression also decreased in the mcmA mutant (TableS2), suggesting that AN4394 functions similarly to EcdR. Regulation of conidial development in A. nidulans is regulated by various transcription factors, and FlbB, FlbC, FlbD are thought to function upstream of BrlA.Citation42) Currently, AN4394 is not well characterized in A. nidulans, however, these Flb proteins might function in concert with McmA for regulation of conidial development.
Consequently, we have shown for the first time in filamentous fungi that the SRF-MADS protein is involved in various cellular functions including cellulase, hemicellulase, and protease production, as well as asexual and sexual development. Identification of partner proteins of McmA in each regulatory system, and also understanding the regulation McmA at the transcriptional and post-transcriptional levels will uncover the precise mechanisms of McmA, a wide domain transcription factor of biological and biotechnological importance.
Author contributions
Tetsuo Kobayashi and Masashi Kato designed the study. Nuo Li, Emi Kunitake, Yoshikazu Endo and Miki Aoyama carried out the experiments. Masashi Kato, Makoto Kimura, and Kyoko Kanamaru supervised all the experiments. Nuo Li drafted the manuscript, and Tetsuo Kobayashi critically revised the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry and by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry.
Supplementary material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2016.1146074
References
- Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21.10.1016/S0378-1119(03)00747-9
- Messenguy F, Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:2586–2592.10.1128/MCB.13.4.2586
- Yoon S, Govind CK, Qiu H, et al. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc. Natl. Acad. Sci. USA. 2004;101:11713–11718.10.1073/pnas.0404652101
- Mead J, Bruning AR, Gill MK, et al. Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol. Cell Biol. 2002;22:4607–4621.10.1128/MCB.22.13.4607-4621.2002
- Maher M, Cong F, Kindelberger D, et al. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell Biol. 1995;15:3129–3137.10.1128/MCB.15.6.3129
- de Nadal E, Casadomé L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell Biol. 2003;23:229–237.10.1128/MCB.23.1.229-237.2003
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057.10.1046/j.1365-2958.1999.01667.x
- Nolting N, Pöggeler S. A MADS box protein interacts with a mating-type protein and is required for fruiting body development in the homothallic ascomycete Sordaria macrospora. Eukaryot. Cell. 2006;5:1043–1056.10.1128/EC.00086-06
- Zhou X, Liu W, Wang C, et al. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol. Microbiol. 2011;80:33–53.10.1111/j.1365-2958.2011.07556.x
- Yamakawa Y, Endo Y, Li N, Yoshizawa M, et al. Regulation of cellulolytic genes by McmA, the SRF-MADS box protein in Aspergillus nidulans. Biochem. Biophys. Res. Commun. 2013;431:777–782.10.1016/j.bbrc.2013.01.031
- Ortiz CS, Shim WB. The role of MADS-box transcription factors in secondary metabolism and sexual development in the maize pathogen Fusarium verticillioides. Microbiol. 2013;159:2259–2268.10.1099/mic.0.068775-0
- Yang E, Chow WN, Wang G, et al. Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei. PLoS genet. 2014;10:e1004662.10.1371/journal.pgen.1004662
- Damveld RA, Arentshorst M, Franken A, et al. The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 2005;58:305–319.10.1111/j.1365-2958.2005.04827.x
- Fujioka T, Mizutani O, Furukawa K, et al. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell. 2007;6:1497–1510.10.1128/EC.00281-06
- Mehrabi R, Ding S, Xu JR. MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot. Cell. 2008;7:791–799.10.1128/EC.00009-08
- Endo Y, Yokoyama M, Morimoto M, et al. Novel promoter sequence required for inductive expression of the Aspergillus nidulans endoglucanase gene eglA. Biosci. Biotechnol. Biochem. 2008;72:312–320.10.1271/bbb.70278
- Rowlands RT, Turner G. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans. Mol. Gen. Genet. 1973;126:201–216.10.1007/BF00267531
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111.10.1093/bioinformatics/btp120
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28:511–515.10.1038/nbt.1621
- Katz ME, Flynn PK, vanKuyk PA, et al. Mutations affecting extracellular protease production in the filamentous fungus Aspergillus nidulans. Mol. Gen. Genet. 1996;250:715–724.
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. New York, NY: Cold Spring Harbor Laboratory Press; 2001.
- Coradetti ST, Xiong Y, Glass NL. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. MicrobiologyOpen. 2013;2:595–609.10.1002/mbo3.94
- Shimizu M, Kaneko Y, Ishihara S, et al. Novel β-1,4-mannanase belonging to a new glycoside hydrolase family in Aspergillus nidulans. J. Biol. Chem. 2015;290:27914–27927.
- vanKuyk PA, Cheetham BF, Katz ME. Analysis of two Aspergillus nidulans genes encoding extracellular proteases. Fun. Genet. Biol. 2000;29:201–210.
- Vienken K, Fischer R. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 2006;61:544–554.10.1111/mmi.2006.61.issue-2
- Vienken K, Scherer M, Fischer R. The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics. 2005;169:619–630.10.1534/genetics.104.030767
- Jin FJ, Nishida M, Hara S, et al. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fun. Genet. Biol. 2011;48:1108–1115.10.1016/j.fgb.2011.10.001
- Emri T, Szilágyi M, László K, et al. PepJ is a new extracellular proteinase of Aspergillus nidulans. Folia Microbiol (Praha). 2009;54:105–109.10.1007/s12223-009-0015-8
- Katz ME, Bernardo SM, Cheetham BF. The interaction of induction, repression and starvation in the regulation of extracellular proteases in Aspergillus nidulans: evidence for a role for CreA in the response to carbon starvation. Curr. Genet. 2008;54:47–55.10.1007/s00294-008-0198-6
- Adams TH, Boylan MT, Timberlake WE. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362.10.1016/0092-8674(88)90198-5
- Hoi JWS, Dumas B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell. 2010;9:480–485.
- Znameroski EA, Li X, Tsai JC, et al. Evidence for transceptor function of cellodextrin transporters in Neurospora crassa. J. Biol. Chem. 2014;289:2610–2619.10.1074/jbc.M113.533273
- Arst HN Jr, Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 1973;126:111–141.10.1007/BF00330988
- Tilburn J, Sarkar S, Widdick DA, et al. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790.
- Punt PJ, Schuren FH, Lehmbeck J, et al. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fun. Genet. Biol. 2008;45:1591–1599.10.1016/j.fgb.2008.09.007
- Cohen BL. Regulation of intracellular and extracellular neutral and alkaline proteases in Aspergillus nidulans. J. Gen. Microbiol. 1973;79:311–320.10.1099/00221287-79-2-311
- Kudla B, Caddick MX, Langdon T, et al. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364.
- Dyer PS, O’Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 2012;36:165–192.10.1111/j.1574-6976.2011.00308.x
- Vallim MA, Miller KY, Miller BL. Aspergillus SteA (Sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 2000;36:290–301.10.1046/j.1365-2958.2000.01874.x
- Primig M, Winkler H, Ammerer G. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 1991;10:4209–4218.
- Nolting N, Pöggeler S. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 2006;62:853–868.10.1111/mmi.2006.62.issue-3
- Yu JH. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology. 2010;38:229–237.10.4489/MYCO.2010.38.4.229