Abstract
Endocytosis is vital for hyphal tip growth in filamentous fungi and is involved in the tip localization of various membrane proteins. To investigate the function of a Wiskott–Aldrich syndrome protein (WASP) in endocytosis of filamentous fungi, we identified a WASP ortholog-encoding gene, wspA, in Aspergillus nidulans and characterized it. The wspA product, WspA, localized to the tips of germ tubes during germination and actin rings in the subapical regions of mature hyphae. wspA is essential for the growth and functioned in the polarity establishment and maintenance during germination of conidia. We also investigated its function in endocytosis and revealed that endocytosis of SynA, a synaptobrevin ortholog that is known to be endocytosed at the subapical regions of hyphal tips in A. nidulans, did not occur when wspA expression was repressed. These results suggest that WspA plays roles in endocytosis at hyphal tips and polarity establishment during germination.
Graphical abstract
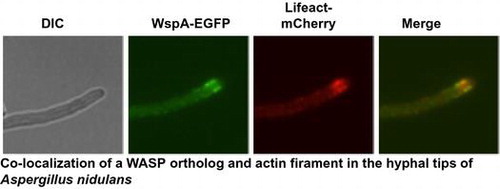
Colocalization of a WASP ortholog and actin filament in the hyphal tips of Aspergillus nidulans.
Polar extension of hyphal tips is a growth characteristic of filamentous fungi. Proper extension depends on both actin filaments and microtubules in the cytoskeleton. In this process, proteins involved in cell wall synthesis and in membranes are supplied by exocytosis and endocytosis occurring in the subapical regions of hyphae.Citation1,2)
Endocytosis is a crucial process for the polar growth of filamentous fungi. In these fungi, many plasma membrane (PM) proteins are endocytosed via small vesicles. These proteins are transported to vacuoles by early and late endosomes (EEs and LEs, respectively) or return through EEs or EEs and Golgi equivalents to fuse with PMs. There are clathrin-mediated endocytic and clathrin-independent endocytic pathways in mammalian cells. In Saccharomyces cerevisiae, however, only the former pathway has been investigated extensively, and approximately, 60 proteins are known to be involved in the process.Citation3,4) Among these proteins, a Wiskott–Aldrich syndrome family protein (WASP) ortholog, Las17p, is known to play a crucial role in this pathway. Since WASPs activate the Arp 2/3 complex, which promotes nucleation of actin filaments, they are also referred to as nucleation-promoting factors (NPFs). Deletion of LAS17 caused aberrant actin filament organization and defects in endocytosis,Citation5,6) and its deletion mutant did not grow at high temperatures. Las17p interacts with G-actin at the C-terminal WH2 domain and with the Arp2/3 complex by its C-terminal acidic sequence. The central proline-rich region of Las17p binds actin and some SH3 domain-containing proteins. This region also has nucleation activity independent of the Arp2/3 complex. Las17p interacts with Vrp1 through the WH1 domain at its N-terminus.Citation4,7–9) Deletion mutants of the genes encoding WASP orthologs in Ashbya gossypii and Candida albicans were defective in polarized growth.Citation10,11) A WASP ortholog-deficient mutant of Cryptococcus neoformans showed defects in cytokinesis, endocytosis, and exocytosis.Citation12)
In filamentous fungi, endocytosis occurs at subapical regions of hyphal tips, where a collar of actin patches is present.Citation13,14) Endocytosis is important to protein and membrane recycling at hyphal apices, and so is essential for the rapid extension of hyphal tips. In Aspergillus nidulans, the gene products of slaB, arfB, fimA, and myoA have been reported to function in endocytosis.Citation15–19) slaB is essential for growth and maintenance of polarity.Citation15,16) Deletion of arfB or fimA has been shown to delay the polarity establishment, resulting in extremely swollen conidia.Citation17,18) In addition, Aoend4, a slaB ortholog of Aspergillus oryzae, also functions in endocytosis, and repression of Aoend4 expression retarded colony growth and caused defects in hyphal morphogenesis.Citation20) myoA, which encodes a class I myosin, is essential for polarized growth.Citation21) A myoA ortholog of A. oryzae, aipB, is suggested to be involved in the endocytosis.Citation22) Collectively, these results demonstrate that endocytosis is required for the establishment and maintenance of polarity in filamentous fungi. The detailed mechanism, however, remains largely unresolved.
In filamentous fungi, some transmembrane proteins at hyphal tips are believed to be endocytosed at subapical regions and transported to apices again during hyphal tip extension. This recycling process keeps these proteins in the hyphal tip regions.Citation23,24)
To clarify the involvement of WASPs in endocytosis and polar growth of filamentous fungi, we identified a WASP ortholog in A. nidulans and characterized its function. We also investigated the effect of depletion of this WASP ortholog on the localization of SynA.
Materials and methods
Strains, media, primers, and transformation
A. nidulans strains used in this study are listed in Table . All strains were grown in minimal medium (MMG) or YG medium.Citation25) Pyridoxine (0.5 mg/mL), uridine (10 mM), and uracil (10 mM) were added when necessary, and each supplement added was indicated by a single lowercase letter after the medium name. To induce the expression of alcA promoter, 100 mM threonine and 0.1% fructose were added to MMG instead of glucose. We referred to the medium as MMTF. Thiamin was added at a final concentration of 100 μM when the expression of thiF promoter was repressed. MMG and YG plates contained 1.5% agar. Strains were grown at 37 °C unless otherwise indicated. Escherichia coli strain DH5α was used for propagation of plasmids. Primers used in this study are listed in Table S1. Bacterial and fungal transformations were performed as described previously.Citation26)
Table 1. A. nidulans strains used in this study.
Isolation of total RNA from A. nidulans
The conidia of the strain A1149 were inoculated into YGuu medium and cultured for 24 h at 37 °C. Mycelial pellets were collected and frozen by liquid nitrogen. Frozen mycelia were ground by a mortar. Isolation of total RNA was done using RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions.
Rapid amplification of cDNA ends (RACE) analysis
RACE analysis was performed using GeneRacer™ Kit for full-length RNA ligase-mediated rapid amplification of 5′ and 3′ cDNA ends (RLM-RACE)(Invitrogen) according to the manufacturer’s instructions.
To confirm the translation initiation site and positions of introns of wspA, we performed RACE analysis using 5-AN11104-rev-GSP and 5-AN11104-rev-GSP-n as a nested primer for 5′ RACE, and 3-AN11104-for-GSP for 3′ RACE.
Colony PCR
Conidia were suspended in 50 μL of extraction buffer (100 mM Tris-HCl pH 9.5, 1 M KCl, and 10 mM EDTA) and heated at 100 °C for 15 min. Then, the solution was vortexing vigorously and centrifuged at 15,000 rpm for 1 min. One μL of the supernatant was used for polymerase chain reactions (PCRs).
Preparation of cell lysate and western blot analysis
Conidia of A. nidulans were inoculated into appropriate liquid medium and incubated for 14–16 h at 37 °C. Mycelial pellets were collected and resuspended in 200 μL of extraction buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA). Then, 1 μL of protease inhibitor cocktail (SIGMA) was added. Cell lysates were prepared with Multi-beads shocker (YASUI KIKAI) as described previously.Citation27) Western blot analysis was performed as described previouslyCitation28) with slight modifications. A mouse anti-GFP antibody (SIGMA) was diluted with Can Get Signal (TOYOBO) at a 1:1000, and used as a primary antibody. A horseradish peroxidase-conjugated rabbit anti mouse IgG antibody (Cell Signaling Technology) was diluted with Can Get Signal at a 1:1000 and used as a secondary antibody.
Constructions of plasmids
A 1.8-kb fragment of BamHI- and SpeI-digested pBSpyrGCitation29) was cloned into BamHI- and SpeI-digested pBluescript II SK+, yielding pBSpyrGII. A 1.0-kb fragment containing the 5′ upstream sequence of thiF ORF was amplified from the total DNA of A1149 using BglII-thiF(p)-f and PstI-thiF(p)-r as primers. The amplified fragment was digested with Bgl II and Pst I and ligated into the BamHI- and PstI-digested pBSpyrGII, yielding ppyrGthiF(p).
A 1.3-kb fragment containing the 5′ upstream sequence of the wspA ORF was amplified from the total DNA of A1149 using 5-(1320)-AN11104-f and 3-AN11104-rev as primers. A 2.7-kb fragment containing pyrG and thiF(p) was amplified from ppyrGthiF(p) using pyrG+1000-481For and thiF(p)-r as primers. A 1.0-kb fragment containing the 5′ region of the wspA ORF was amplified from the total DNA of A1149 using thiF(p)-AN11104-f and 5-AN11104-rev-GSP as primers. The 1.3-kb, 2.7-kb, and 1.0-kb fragments described above were assembled by fusion PCRCitation30) using XbaI-5-AN11104-f and PstI-AN11104-mid-r as primers. The 5.0-kb assembled fragment was digested with XbaI and PstI and ligated into XbaI- and PstI-digested pUC118, yielding pthiF(p)-AN11104.
A 2.1-kb fragment containing the entire ORF of wspA was amplified from the total DNA of A1149 using AN11104-for and egfp-AN11104-rev as primers. A 0.7-kb fragment containing the gene encoding EGFP was amplified from pEGFP (Clontech) using egfp-for and egfp-rev as primers. A 0.5-kb fragment containing 3′ downstream sequence of the wspA ORF was amplified from the total DNA of A1149 using egfp-AN11104ter-for and pyroA-AN11104ter-rev as primers. The 2.1, 0.7, and 0.5-kb amplified fragments were assembled by the fusion PCR using AN11104-for and pyroA-AN11104ter-rev as primers, yielding a 3.3-kb fragment.
A 2.4-kb fragment containing the entire pyroA gene was amplified from pUCpyroA2Citation31) using pyroA-5n and pyroA-3n as primers. The 3.3-kb fragment and the 2.4-kb fragment were assembled using AN11104-nest-for and pyroA-nest-rev as primers, yielding a 5.7-kb fragment. A 2.9-kb fragment containing the 3′ part of wspA ORF and the 3′ downstream sequence was amplified from the total DNA of A1149 using pyroA-mid-AN11104-for and 3-AN11104-2kb-rev as primers. A 2.4-kb fragment containing the entire pyroA gene was amplified from pUCpyroA2 using pyroA-1st-for and pyroA-3n as primers and assembled with the 2.9-kb fragment using pyroA-5n and 3-AN11104-2kb-n-rev, yielding a 5.3-kb fragment. A 3.6-kb fragment produced by BamHI-SalI digestion of the 5.7-kb fragment and a 3.4-kb fragment produced by BamHI-SalI digestion of the 5.3-kb fragment were ligated into BamHI-diegested pUC118, yielding pAN11104-EGFP.
pACLAsh was constructed as follows: A 8.1-kb fragment was amplified from ppyrGLACitation29) using 3EGFPR and mC-pyrGR as primers. A 0.7-kb fragment containing mCherry-coding region was amplified from pmCherry-N1 (Clontech) using SmaI-mCherry-f and mCherry-r as primers. The 8.1-kb and 0.7-kb fragments were connected and amplified by fusion PCR using Link2R and SmaI-mCherry-f as primers. The amplified fragment was digested with SmaI and self-ligated, yielding ppGALLAmC. A 7.1-kb fragment was amplified from ppGALLAmC using abp140F and pyrGdown-Nde as primers. A 1.0-kb fragment containing actA promoter was amplified from A1149 total DNA using SmaI-actA(p)F and LA-actA(p)R as primers. The 7.1-kb and 1.0-kb fragments were connected and amplified by fusion PCR using pyrGdown-Nde and SmaI-actA(p)F as primers. The amplified fragment was digested with SmaI and self-ligated, yielding ppGACLAmC. The 3.3-kb SpeI-fragment of ppGACLAmC containing actA(p), Lifeact-mCherry, and pyrG was ligated with SpeI-digested pBSII, yielding pACLAsh.
A1.4-kb fragment containing 0.5–1.9 kb upstream of synA was amplified from the total DNA of A1149 using EcoRI-5-synA-f and pyroA-5-synA-r. A 2.4-kb fragment containing the entire pyroA gene was amplified from pUCpyroA2 using pyroA5n and pyroA3n as primers. These two fragments were fused by the fusion PCR using EcoRI-5-synA-f and pyroA3n as primers, yielding a 3.8-kb fragment. A 0.5-kb fragment of 5′ upstream of synA was amplified using pyroA-synA(p)-f and EGFP-synA(p)-r as primers. A 0.7-kb fragment containing egfp was amplified from pEGFP using egfp-for and egfp-rev as primers. A 1.0-kb fragment containing the entire synA ORF and its 0.4-kb downstream region was amplified from the total DNA of A1149 using EGFP-synA-f and synA-r as primers. The 0.5-kb, 0.7-kb, and 1.0-kb fragments amplified above were fused by the fusion PCR using pyroA-synA(p)-f and synA-r as primers, yielding a 2.2-kb fragment. This 2.2-kb fragment and the 2.4-kb fragment containing pyroA were fused by the fusion PCR using pyroA5n and synA-r-n as primers, yielding a 4.6-kb fragment. This 4.6-kb fragment was digested with MluI and SmaI and the 3.8-kb fragment containing 5′ upstream of synA and the entire pyroA gene was digested with EcoRI and MluI. These two fragments were ligated into EcoRI- and SmaI-digested pUC18, yielding pEGFP-SynA.
Constructions of strains
To obtain a wspA deletion mutant, we constructed the strain as follows: A 1.0-kb fragment containing 5′ upstream sequence of the wspA ORF was amplified from the total DNA of AN1149 using 5-AN11104-for and 5-AN11104-rev as primers. A 2.0-kb fragment containing the entire pyrG gene was amplified from pPyrG+750 containing the entire coding region and 715 bp upstream of pyrG (Ono-Tanaka et al. unpublished) using pyrG+1000-481For and pyrG+1000-2480Rev as primers. A 1.9-kb fragment containing the 3′ region and the downstream sequences of the wspA ORF was amplified from the total DNA of AN1149 using mid-AN11104-for and 3-AN11104-rev as primers. These three amplified fragments were assembled by fusion PCR using 5-AN11104-nest-for and 3-AN11104-nest-rev as primers, yielding a 4.9-kb fragment. A1149 was transformed with the 4.9-kb fragment, yielding heterokaryons wspA-hk-1 and -2.
The strain that produces WspA-EGFP was constructed as follows: AN1149 was transformed with the 7.0-kb fragment obtained from the BamHI-digested pAN11104-EGFP. Two transformants were selected in the transformation. The replacement of the wild-type wspA in these transformants with wspA-egfp was confirmed by Southern blot analysis. We designated these transformants as wspA-EGFP-1 and -2.
wspA-EGFP-1 was transformed with BlnI-digested pACLAsh. A transformant in which actA(p)-lifeact-mcherry was integrated into the pyrG locus was selected by Southern blot analysis and designated it wspA-EGFP/actLAmCh.
The strains that produce WspA under the control of the thiF promoter were constructed as follows: A1149 was transformed with a 4.5-kb fragment obtained from the EcoRV- and SpeI-digested pthiF(p)-AN11104. Two transformants were selected in the transformation. The replacement of the wild-type wspA in these transformants with the thiF(p)-wspA was confirmed by Southern blot analysis. We designated these transformants as thiF(p)-wspA-1, and -2.
A1149 and thiF(p)-wspA were transformed with a 5.6-kb fragment obtained from EcoRI- and SmaI-digested pEGFP-SynA. Two transformants were selected in each transformation and the production of EGFP-SynA was confirmed by western blot analysis using anti-GFP antibody. We designated these strains, EGFP-synA-1, and -2, and thiF(p)-wspA/EGFP-synA-1 and -2, respectively.
All the strains described above that have the same genotypes exhibited the same phenotypes under the conditions tested.
A1149 was transformed with a 2.7-kb BamHI-PstI fragment of pUCPYROA.Citation32) A transformant in which pyroA4 mutation was replaced with the wild-type pyroA was designated A1149/pyroA.
Uptake of FM4-64
Uptake of FM4–64 was performed as follows: thiF(p)-wspA and A1149/pyrG-1 were inoculated in MMGp with or without 100 μM thiamin and incubated for 8 h at 37 °C in glass-based dishes. Then, the dishes were cooled on ice for 10 min and the medium was removed. The fresh medium containing 10 μM FM4-64 was added and incubated for 10 min on ice. Then, the dishes were washed 3 times with fresh medium and were incubated at 25 °C.
Fluorescence microscopy
Calcofluor white (CFW) staining and fluorescence microscopy was done as previously described.Citation25) Conforcal laser microscopy was done using Fluoview FV500 system (Olympus).
Results
Characterization of a LAS17 ortholog in A. nidulans
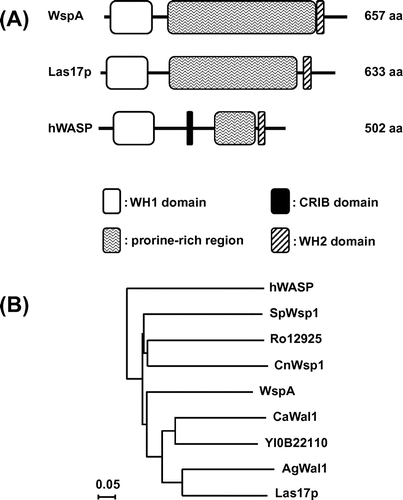
There is an ortholog of S. cerevisiae LAS17, AN11104, in the genome of A. nidulans (http://www.aspgd.org). A 5′ and 3′ RACE analysis showed that this gene contains two introns and encodes a protein of 657 amino acids. The positions of introns are shown in Fig. S1. The product of AN11104 exhibits 75 and 62% amino acid similarities with Las17p and human WASP, respectively, and they have similar domain organizations, containing WH (WASP homology) 1, proline-rich, and WH2. Thus, we refer to AN11104 as wspA (Fig. (A)). The wspA gene product, WspA, has a sequence rich in acidic amino acids at its C-terminus, similar to other WASP orthologs (Fig. S2). However, a CRIB domain, which is located in the central region of a human WASP and is known to bind Cdc42, is not present in Las17p or WspA. A phylogenetic tree of WASP orthologs is shown in Fig. (B).
Fig. 1. Domain organizations (A) and a phylogenetic tree (B) of the WspA orthologs.
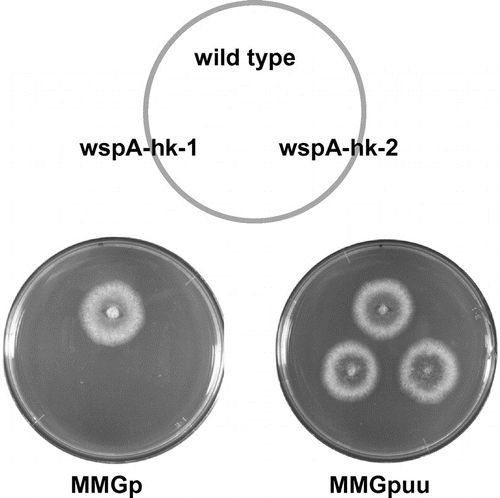
To investigate the function of wspA, we attempted to construct its deletion mutant (see Materials and methods and strategy is shown in Fig. (A)). Two transformants grew poorly on the selective medium, MMGp, and formed small colonies (Fig. (B)). We designated these transformants as wspA-hk-1 and wspA-hk-2 and analyzed the integration loci of the transformed DNA in their genomes by colony PCR (Fig. (C)). When P2 and P3 were used as primers, one fragment was amplified from the wspA-hk-1 and wspA-hk-2 DNAs, whereas no fragment was amplified from the A1149/pyrG-1 (Fig. (C) lanes 2–4). When P1 and P2 were used as primers, two fragments (indicated by arrows) were amplified from the wspA-hk-1 and wspA-hk-2 DNAs, whereas one fragment was amplified from the A1149/pyrG-1 DNA (Fig. (C) lanes 5–7). These results indicated that wspA-hk-1 and wspA-hk-2 are heterokaryons. Since the conidia of A. nidulans contain one nucleus, there should be two types of the conidia in these transformants that have either a ΔwspA pyrG+ nucleus or a wspA+ pyrG- nucleus. Their conidia did not form colonies when inoculated on the selective MMGp medium, at 37 °C, whereas they grew well on the nonselection MMGpuu medium (Fig. ). The conidia of the heterokaryons did not form colonies on the selective medium at 25 °C or 30 °C (data not shown). These results suggest that wspA is essential for the growth of A. nidulans. The growth defect of these heterokaryons is likely caused by the insufficiency of the WspA protein production in the hyphae.
Fig. 2. Characterizations of wspA-hk-1 and -2.
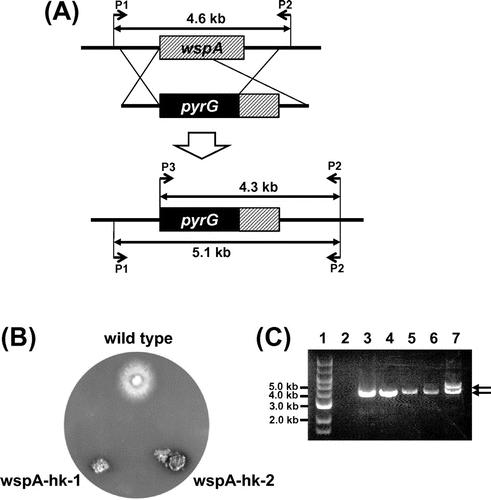
Fig. 3. Growth of wspA-hk-1 and wspA-hk-2 on the selective and non-selective media.
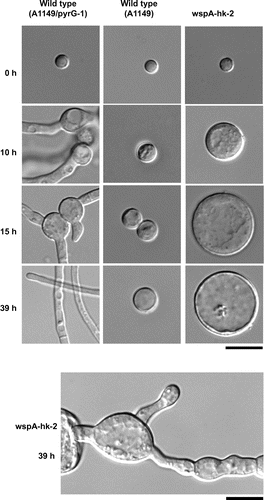
To determine the terminal phenotype of the wspA deletion mutant, the conidia of wspA-hk-2 were inoculated into the selective liquid medium and incubated for 39 h. Some conidia swelled slightly and stopped growing. This phenotype is similar to that of a pyrG- mutant (Fig. , A1149), suggesting that these conidia contained a wspA+ pyrG- nucleus. In contrast, other conidia swelled more and did not form germ tubes even after 39 h of incubation at 37 °C. Occasionally, swollen conidia with germ tubes were also observed (Fig. , lower panel). However, these germ tubes did not grow further and did not form colonies. When conidia of wspA-hk-2 were incubated in the selective medium at 25 °C, some conidia continued to grow isotropically and, after 115 h of incubation, formed extremely large cells (Fig. S3). These results suggest that the swollen conidia contained ΔwspA pyrG+ nuclei and that wspA plays a crucial role in the polarity establishment and/or maintenance during conidial germination.
Fig. 4. Terminal phenotypes of the wspA deletion mutant.
Localization of WspA in the hyphae
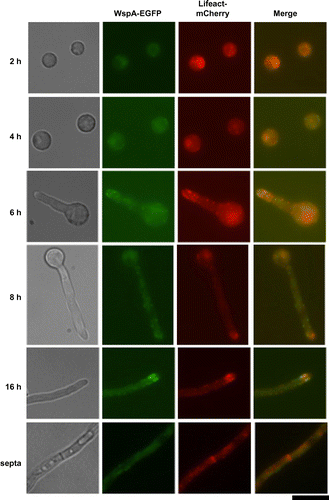
Localization of WspA in the germinating conidia and growing hyphae was analyzed using WspA tagged with EGFP at its C-terminus. The wspA-EGFP strains produced WspA-EGFP (Fig. S4(A)) and grew as well as the wild-type strain (Fig. S4(B) and data not shown). The calculated molecular mass of WspA-EGFP was 94 kDa, whereas the mass determined by SDS-PAGE was approximately 130 kDa (Fig. S4(A)). This suggests that WspA is modified in A. nidulans. Since WASP orthologs are known to interact with actin, we constructed the strain, wspA-EGFP/actLAmCh, which simultaneously produces WspA-EGFP and Lifeact tagged with mCherry (see Materials and methods). Lifeact consists of 17 N-terminal amino acids of Abp140p in S. cerevisiae, and it is used to observe the localization of F-actin in Neurospora crassa and A. nidulans.Citation29,33) The wspA-EGFP/actLAmCh strain grew as well as the wild-type strain (Fig. S4(B)), indicating that WspA-EGFP and Lifeact-mCherry are functional.
During conidial germination, WspA-EGFP localized to the cortex of swelling conidia in patches before germ tube formation. WspA-EGFP and Lifeact-mCherry did not colocalized at that time (Fig. , 4 h). After germ tube formation, however, WspA-EGFP primarily localized to the tips of germlings, appearing as punctate structures that partially colocalized with those of Lifeact-mCherry (Fig. , 6 and 8 h). WspA-EGFP was mainly observed at the cortexes of subapical regions of hyphae, where it colocalized with Lifeact-mCherry in mature hyphae (Fig. , 16 h). These results suggest that WspA is colocalized with actin filaments at the subapical regions where endocytosis occurs. WspA-EGFP was not observed at septa (Fig. , septa).
Fig. 5. Co-localization of WspA and F-actin in A. nidulans.
The distributions of WspA-EGFP in the wspA-EGFP strain were nearly the same as those in the wspA-EGFP/actLAmCh strain, appearing in swelled conidia, germlings, and mature hyphae (data not shown), suggesting that the expression of Lifeact-mCherry did not disturb the localization of WspA-EGFP.
Function of WspA in the endocytosis
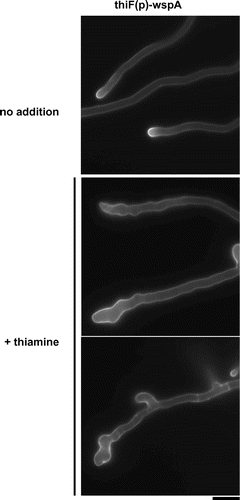
To investigate the function of WspA in endocytosis, we constructed a conditional mutant of wspA, the thiF(p)-wspA strain, which expresses wspA under the control of a thiF promoter. Expression of the thiF promoter is repressed when thiamine is added to the medium.Citation34) The thiF(p)-wspA strain grew as well as the wild-type strain under the thiF(p)-inducing condition, while growth of the thiF(p)-wspA strain was, markedly retarded under the thiF(p)-repressing condition (Fig. S5). Growth of the thiF(p)-wspA strain is likely a result of leaky expression of wspA under the thiF(p)-repressing condition. It was not restored when 1.2 M sorbitol was added to the medium (Fig. S5). The hyphae of the strain were highly branched, and their lateral walls were not smooth under the thiF(p)-repressing condition (Fig. ).
Fig. 6. Morphology of the hyphae under the wspA-repressing condition.
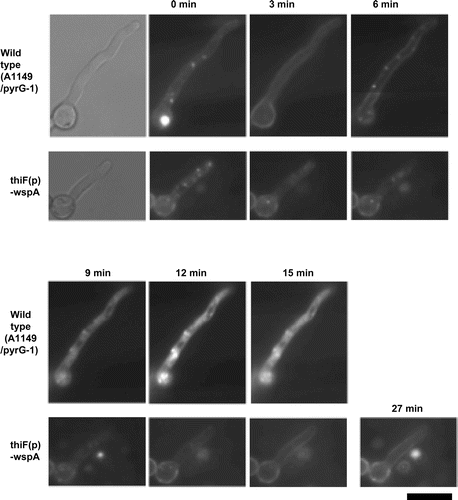
To investigate the involvement of WspA in endocytosis, we observed the internalization of FM4-64 in the thiF(p)-wspA strain (Fig. ). FM4-64 is a lipophilic fluorescent dye frequently used to monitor endocytosis. When cells are treated with FM4-64, it binds PM and is subsequently internalized by endocytosis. Finally, it is transported to the vacuole. Under the thiF(p)-repressing condition, FM4-64 was internalized after 6 min of incubation at 37 °C and reached to organelles after 9 min in the wild-type strain, while, in the thiF(p)-wspA strain, only the cell surface was stained after 27 min of incubation (Fig. ). In contrast, under the thiF(p)-inducing condition, internalization was observed in both strains after 6 min of incubation (Fig. S6). These results suggest that WspA is involved in endocytosis.
Fig. 7. Internalization of FM4-64 in the thiF(p)-wspA strain under the wspA-repressing conidition.
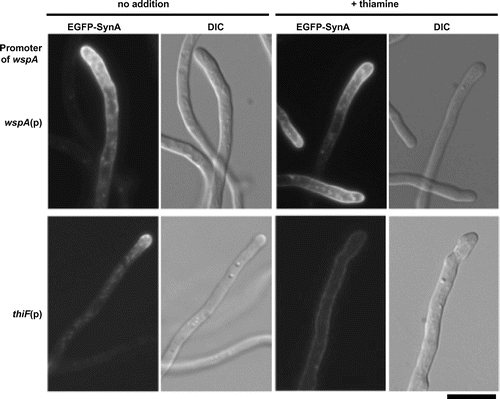
Next, we used an endocytic marker protein, SynA, to monitor endocytosis. SynA is a synaptobrevin ortholog of A. nidulans that localizes at hyphal tips. SynA is known to be endocytosed from subapical regions of hyphal tips, and it is recycled to the extreme apices of hyphae.Citation13,14) We constructed a thiF(p)-wspA/EGFP-synA strain, a wspA conditional mutant that produces EGFP-SynA, and examined the localization of EGFP-SynA. In the thiF(p)-wspA/EGFP-synA strain, EGFP-SynA predominantly localized at hyphal tips under the thiF(p)-inducing condition (Fig. , no addition), whereas it was distributed to the surface of basal regions of hyphae under the thiF(p)-repressing condition (Fig. , + thiamine). In the wild-type strain, the distribution of EGFP-SynA was similar in both conditions. Collectively, the results indicate that WspA functions in endocytosis at the subapical regions.
Discussion
Endocytosis is crucial for hyphal tip growth in filamentous fungi and the tip localization of certain transmembrane proteins. WASPs are known to be involved in the endocytosis in mammals and yeasts. To investigate the function of the WASP ortholog in filamentous fungi, we identified a WASP ortholog-encoding gene of A. nidulans, wspA, and characterized it. wspA is essential for the growth of the organism and plays an important role in the establishment and/or maintenance of polarity during conidial germination and hyphal tip growth. We showed that WspA predominantly localized at the tips of germ tubes and subapical regions near the actin ring-like structures of mature hyphae. Repression of wspA expression reduced the incorporation of FM4-64 and changed the tip localization of SynA, indicating that WspA functions in endocytosis.
WspA consists of a WH1 domain, a proline-rich region, a WH2 domain, and a C-terminal acidic region. The C-terminal acidic region is known to be a binding site for the Arp2/3 complex, and it is crucial for the activation of the Arp2/3 complex to promote actin filament nucleation in yeast and mammalian cells.Citation7,35) Polyproline tracts in the proline-rich region of Las17p and mammalian WASP also bind to actin and have NPF activity independent of the Arp2/3 complex.Citation7) Since polyproline tracts in the proline-rich region and the C-terminal acidic region are both conserved in WspA, it is probable that WspA is also involved in actin filament nucleation in Arp2/3-dependent and Arp2/3-independent manners.
In filamentous fungi, filamentous actin plays crucial roles in hyphal tip growth by delivering exocytic vesicles to the apical PM and endocytic recycling at subapical regions. WspA colocalized with actin filament at the subapical regions of mature hyphae. In S. cerevisiae, Las17p was reported to form a large stable complex with Sla1p. This complex may be recruited to endocytic sites before actin polymerization.Citation36) The polyproline tract of Las17p binds to the SH3 regions of Sla1p to form the complex. A SLA1 ortholog gene, AN1462, is present in the genome of A. nidulans and its gene product, AN1462, is predicted to have 4 SH3 domains. Thus, it is possible that WspA is also recruited to subapical regions of hyphal tips with AN1462 and regulates endocytosis there through the nucleation of actin filaments.
wspA is essential for the growth of A. nidulans at 25, 30, and 37 °C. In S. cerevisiae, a deletion mutant of las17 grew slowly at 30 °C and did not grow above 34 °C.Citation5) Temperature sensitive growth phenotypes were also observed in deletion mutants of wspA orthologs in Schizosaccharomyces pombe,Citation37) A. gossypii,Citation10) and C. neoformans.Citation12) In contrast, the deletion mutant of a wspA ortholog in C. albicans did not show temperature sensitivity, but exhibited defects in filamentous growth.Citation11) These results suggest that the functional contributions of these WASP orthologs to growth are somewhat different in these organisms. Among these deletion mutants, the mutants of A. gossypii and C. albicans showed defects in polarized growth, suggesting that WASP proteins are crucial for the polarized growth. The fact that wspA is essential for the growth supports this possibility.
In the wspA deletion mutant of A. nidulans, germ tubes hardly formed, indicating that the polarity establishment during germination is severely deficient (Fig. ). Defects in the maintenance of polarity were observed in the deletion mutants of endocytosis-related genes, slaB, arfB, and fimA in A. nidulans.Citation15,17,18,21) Most conidia of the wspA deletion mutants did not form germ tubes, suggesting that WspA plays an important role in polarity establishment during germination. However, portions of conidia did form germ tubes. This indicates that the function of WspA is not essential for the polarity establishment. It is probable that WspA also plays a role in maintaining polarity because the germ tubes formed by the deletion mutant were abnormally thick, with aberrant morphologies.
In yeasts, WASP-orthologs form complexes with verprolin and recruit type-I myosins at actin patches. There, they function together in endocytosis.Citation38–40) In A. nidulans, there is only one type-I myosin, MyoA, which is essential for growth and plays a role in endocytosis.Citation19,21,41) MyoA localizes primarily at hyphal tips and forming septa,Citation42) where actin filaments also localize. Since an ortholog of verprolin-encoding genes in yeasts is present in the genome of A. nidulans, it is probable that the mechanism of type-I myosin recruitment at sites of endocytosis is conserved between yeasts and A. nidulans. Type-I myosins possess both NPF activity and motor activity, and these activities are separable.Citation38) In S. cerevisiae, the NPF activities of Las17p and type-I myosins are redundant, and either of them is sufficient for endocytosis to proceed.Citation39,43,44) Since the motor activity of MyoA is suggested to be nonessential for the growth,Citation41) it is possible that MyoA NPF activity is essential for the growth of A. nidulans. This may suggest that the NPF activities of WspA and MyoA are both required for endocytosis in A. nidulans. Further, functional analyses of each domain of WspA and MyoA will clarify specific roles of WASPs and type-I myosins in endocytosis in filamentous fungi.
Authors contribution
H. Horiuchi conceived and designed the experiments; H. Hoshi, LZ, and H. Horiuchi performed the experiments; H. Hoshi, LZ, and H. Horiuchi analyzed the data; H. Horiuchi and AO contributed reagents/materials/analysis tools; H. Horiuchi wrote the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental materials
The supplemental material for this paper is available at http://dx.doi.org/10.1080/23311843.2016.1148580.
suppl_data.zip
Download Zip (1.3 MB)Acknowledgment
We thank H. Yamazaki and T. Katayama for constructing plasmids. This work was done using the facilities of the Biotechnology Research Center, The University of Tokyo.
Notes
Abbreviations: WASP, Wiskott-Aldrich syndrome protein; EGFP, enhanced green fluorescent protein.
References
- Takeshita N, Manck R, Grün N, et al. Interdependence of the actin and the microtubule cytoskeleton during fungal growth. Curr. Opin. Microbiol. 2014;20:34–41. 10.1016/j.mib.2014.04.005
- Schultzhaus Z, Shaw B. Endocytosis and exocytosis in hyphal growth. Fungal Biol. Rev. 2015;29:43–53.10.1016/j.fbr.2015.04.002
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13.10.1016/j.tcb.2011.09.001
- Goode BL, Eskin JA, Wendland B. Actin and endocytosis in budding yeast. Genetics. 2015;199:315–358.10.1534/genetics.112.145540
- Li R. Bee1, a yeast protein with homology to wiscott-aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 1997;136:649–658.10.1083/jcb.136.3.649
- Naqvi SN, Zahn R, Mitchell DA, et al. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 1998;8:959–962.10.1016/S0960-9822(98)70396-3
- Urbanek AN, Smith AP, Allwood EG, et al. A novel actin-binding motif in Las17/WASP nucleates actin filaments independently of Arp2/3. Curr. Biol. 2013;23:196–203.10.1016/j.cub.2012.12.024
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 1999;9:501–505.10.1016/S0960-9822(99)80218-8
- Feliciano D, Tolsma TO, Farrell KB, et al. A second Las17 monomeric actin-binding motif functions in Arp2/3-dependent actin polymerization during endocytosis. Traffic. 2015;16:379–397.10.1111/tra.12259
- Walther A, Wendland J. Apical localization of actin patches and vacuolar dynamics in Ashbya gossypii depend on the WASP homolog Wal1p. J. Cell Sci. 2004;117:4947–4958.10.1242/jcs.01377
- Walther A, Wendland J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich syndrome protein homolog Wal1p. Eukaryotic Cell. 2004;3:471–482.10.1128/EC.3.2.471-482.2004
- Shen G, Whittington A, Wang P. Wsp1, a GBD/CRIB domain-containing WASP homolog, is required for growth, morphogenesis, and virulence of Cryptococcus neoformans. Eukaryotic Cell. 2011;10:521–529.10.1128/EC.00274-10
- Abenza JF, Pantazopoulou A, Rodríguez JM, et al. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75.10.1111/j.1600-0854.2008.00848.x
- Taheri-Talesh N, Horio T, Araujo-Bazan L, et al. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell. 2008;19:1439–1449.10.1091/mbc.E07-05-0464
- Araujo-Bazán L, Peñalva MA, Espeso EA. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol. Microbiol. 2008;67:891–905.10.1111/mmi.2008.67.issue-4
- Hervas-Aguilar A, Penalva MA. Endocytic machinery protein SlaB Is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans. Eukaryot Cell. 2010;9:1504–1518.10.1128/EC.00119-10
- Upadhyay S, Shaw BD. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol. Microbiol. 2008;68:690–705.10.1111/j.1365-2958.2008.06178.x
- Lee SC, Schmidtke SN, Dangott LJ, et al. Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot. Cell. 2008;7:1278–1288.10.1128/EC.00039-08
- Yamashita RA, May GS. Constitutive activation of endocytosis by mutation of myoA, the myosin I gene of Aspergillus nidulans. J. Biol. Chem. 1998;273:14644–14648.10.1074/jbc.273.23.14644
- Higuchi Y, Shoji JY, Arioka M, et al. Endocytosis is crucial for cell polarity and apical membrane recycling in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell. 2009;8:37–46.10.1128/EC.00207-08
- McGoldrick CA, Gruver C, May GS. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J. Cell Biol. 1995;128:577–587.10.1083/jcb.128.4.577
- Matsuo K, Higuchi Y, Kikuma T, et al. Functional analysis of Abp1p-interacting proteins involved in endocytosis of the MCC component in Aspergillus oryzae. Fungal Genet. Biol. 2013;56:125–134.10.1016/j.fgb.2013.03.007
- Shaw BD, Chung DW, Wang CL, et al. A role for endocytic recycling in hyphal growth. Fungal Biol. 2011;115:541–546.10.1016/j.funbio.2011.02.010
- Peñalva MA. Endocytosis in filamentous fungi: Cinderella gets her reward. Curr. Opin. Microbiol. 2010;13:684–692.10.1016/j.mib.2010.09.005
- Fukuda K, Yamada K, Deoka K, et al. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot. cell. 2009;8:945–956.10.1128/EC.00326-08
- Tsuizaki M, Ohta A, Horiuchi H. Myosin motor-like domain of class VI chitin synthase CsmB of Aspergillus nidulans is not functionally equivalent to that of class V chitin synthase CsmA. Biosci. Biotechnol. Biochem. 2013;77:369–374.10.1271/bbb.120822
- Takeshita N, Ohta A, Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell. 2005;16:1961–1970.10.1091/mbc.E04-09-0761
- Takeshita N, Ohta A, Horiuchi H. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide, and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 2002;298:103–109.10.1016/S0006-291X(02)02418-X
- Katayama T, Uchida H, Ohta A, et al. Involvement of protein kinase C in the suppression of apoptosis and in polarity establishment in Aspergillus nidulans under conditions of heat stress. PLoS One. 2012;7:e50503.10.1371/journal.pone.0050503
- Szewczyk E, Nayak T, Oakley CE, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3120.
- Katayama T, Ohta A, Horiuchi H. Protein kinase C regulates the expression of cell wall-related genes in RlmA-dependent and independent manners in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2015;79:321–330.10.1080/09168451.2014.973365
- Ichinomiya M, Yamada E, Yamashita S, et al. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell. 2005;4:1125–1136.10.1128/EC.4.6.1125-1136.2005
- Lichius A, Berepiki A, Read ND. Form follows function – the versatile fungal cytoskeleton. Fungal Biol. 2011;115:518–540.10.1016/j.funbio.2011.02.014
- Calcagno-Pizarelli AM, Hervas-Aguilar A, Galindo A, et al. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J. Cell Sci. 2011;124:4064–4076.10.1242/jcs.088344
- Padrick SB, Rosen MK. Physical Mechanisms of Signal Integration by WASP Family Proteins. Annu. Rev. Biochem. 2010;79:707–735.10.1146/annurev.biochem.77.060407.135452
- Feliciano D, Di Pietro SM. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol. Biol. Cell. 2012;23:4256–4272.10.1091/mbc.E11-12-1022
- Hayles J, Wood V, Jeffery L, et al. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053.10.1098/rsob.130053
- Sun YD, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 2006;11:33–46.10.1016/j.devcel.2006.05.008
- Evangelista M, Klebl BM, Tong AHY, et al. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 2000;148:353–362.10.1083/jcb.148.2.353
- Sirotkin V, Beltzner CC, Marchand JB, et al. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 2005;170:637–648.10.1083/jcb.200502053
- Liu X, Osherov N, Yamashita R, et al. Myosin I mutants with only 1% of wild-type actin-activated MgATPase activity retain essential in vivo function(s). Proc. Natl. Acad. Sci. USA. 2001;98:9122–9127.10.1073/pnas.161285698
- Yamashita RA, Osherov N, May GS. Localization of wild type and mutant class I myosin proteins in Aspergillus nidulans using GFP-fusion proteins. Cell Motil. Cytoskel. 2000;45:163–172.10.1002/(ISSN)1097-0169
- Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 2000;148:363–374.10.1083/jcb.148.2.363
- Lewellyn EB, Pedersen RTA, Hong J, et al. An engineered minimal WASP-myosin fusion protein reveals essential functions for endocytosis. Dev. Cell. 2015;35:281–294.10.1016/j.devcel.2015.10.007