Abstract
Transmembrane protein CD36 is considered to bind its distinct ligands such as long-chain fatty acids primarily by recognizing their terminal carboxyl moiety. In this study, we provide evidence that long-chain fatty aldehydes, such as oleic aldehyde, can be recognized by CD36. We suggest that a single aldehyde group may also serve as one of the structural elements recognizable by CD36.
CD36 is a member of the class B scavenger receptors characterized by two transmembrane domains, a broad expression pattern, and an ability to recognize a variety of ligands.Citation1) The primary function of CD36 is considered to participate in the capture (i.e. clearance) of modified lipoproteins and advanced glycation products that would cause damage to tissues.Citation1) One of the most well-studied CD36 ligands is oxidized low-density lipoprotein (oxLDL).Citation1–3) In an earlier study, the particles were found to undergo capture by the receptor protein because of the surface localization of distinct forms of oxidized phosphatidylcholine (oxPC) (referred to as CD36-specific oxPC ligands or oxPCCD36), which have a terminal γ-hydroxyl(or oxo)-α,β-unsaturated carbonyl on the sn-2 acyl group.Citation2) The terminal moiety was postulated to be essential for recognition by CD36, because non-oxidized counterparts of oxPCCD36 exhibited no detectable interaction with the receptor.Citation2) Later, by a study with a series of artificial constructs of oxPC, a terminal negatively charged carboxylate at sn-2 position was demonstrated to serve as an important requisite for binding with high affinity to CD36.Citation3)
CD36 is also believed to participate in the cellular uptake, trafficking, and metabolism of long-chain fatty acids (LCFAs).Citation4) It would not be surprising if LCFAs should be directly recognized by CD36, because the lipids have a terminal carboxyl group. We found recently that (i) fluorescently labeled oxLDL (fl-oxLDL) bound specifically and saturably to a chemically synthesized peptide representing the oxLDL-binding site on mouse CD36 (amino acid 150 to 168) (hereinafter CD36 mimic) and (ii) the binding of fl-oxLDL to the mimic was inhibited by unsaturated LCFAs, such as the oleic and linoleic acids, as well as by an oxPCCD36 with a terminal carboxyl group at sn-2 position.Citation5) These data provided one piece of molecular evidence that distinct LCFAs could indeed be recognized by CD36. On the other hand, fatty acid derivatives, oleic alcohol, and methyl oleate had no inhibitory effects.Citation5) Based on others’ and our findings,Citation2,3,5) one can propose that a terminal carboxyl group linked to (or concomitant with) distinct hydrocarbon chains could serve as a structural element recognized by CD36.
Our aim was to identify potential structural elements recognizable by CD36, which correspond to the carboxyl group in LCFAs. For this purpose, we attempted to examine the ability of lipids with only one distinct species of hydrocarbon chain to inhibit the binding of fl-oxLDL to a CD36 mimic, named herein CD36150–168 (a 22-amino acid peptide with an N-terminal biotinCitation5)). If certain of test lipids exhibited an inhibitory effect, the acceptability of the non-hydrocarbon chain moieties by CD36 would be indicated. We have previously shown that the degree of inhibition of fl-oxLDL binding to CD36150–168 varies among unsaturated LCFAs.Citation5) Thus, the hydrocarbon chain of test lipids should be identical. In this study, we selected oleic acid as a reference standard and several lipids with oleyl group(s) [CH3(CH2)7CH=CH(CH2)7] as test lipids, based on the criteria that they are present in the diets of human (or possibly those of other distinct mammals) or occur in their gastrointestinal tracts as digests.Citation6–8)
The purity of test lipids obtained from the manufacturers was at least more than 95%. Cholesteryl oleate, cis-11-hexadecenal, 1,2-dioleoyl-rac-glycerol (1,2-diolein), oleamide, oleic acid, oleyl (oleic) alcohol were from Sigma-Aldrich Japan (Tokyo). Trioctadecenoin (triolein) and 1,3-diolein were from Nu-Chek Prep (Elysian, MN). Monoacylglycerols (1-monoolein and 2-monoolein) were from Olbracht Serdary Research Laboratories (Toronto, Canada). Octadecanal (stearic aldehyde) was from Tokyo Chemical Industry (Tokyo). 4-Hydroxy nonenal (4-hydroxy-trans-2-nonenal) dissolved in ethanol at a concentration of 1% w/v was from Cayman Chemical (Ann Arbor, MI). Oleic aldehyde (also referred to as cis-9-octadecenal) was prepared by the chemical conversion of oleic acid. Briefly, oleic acid was once converted into oleic alcohol by treatment with lithium aluminum hydride. The alcohol obtained was purified by silica gel column chromatography and then converted into oleic aldehyde by treatment with 2-iodoxybenzoic acid. The product was purified by silica gel column chromatography and subjected to 1H- and 13C-nuclear magnetic resonance spectrometries and gas chromatograph–mass spectrometry for quality assessment. The purity of oleic aldehyde we obtained was more than 98%. Each of the test lipids (except cholesteryl oleate) was dissolved in ethanol (99.5% v/v, Nacalai Tesque, Kyoto, Japan) at a concentration of less than 3% w/v and stored at −20°C until use. Five milligrams of cholesteryl oleate was suspended in 0.77 mL of ethanol and stored at −20°C until use. The procedures for preparation of fl-oxLDL and CD36150–168 and the protocol for fl-oxLDL binding and competition assay using the CD36 mimic were exactly the same as described previously.Citation5) Briefly described for the assay protocol, the wells of a streptavidin-coated 96-well plate were prewashed with phosphate-buffered saline (PBS, 8 mM Na2HPO4, 1.5 mM KH2PO4, 136 mM NaCl, and 2.7 mM KCl, pH 7.4) with 0.4% w/v bovine serum albumin (BSA) (PBS–BSA). PBS–BSA alone or PBS–BSA with CD36150–168 at a concentration of 5 µM was added to each of the wells. Then, the plate was incubated for 1 h at ambient temperature. After washing with PBS with 0.05% w/v BSA, each well received assay mixture (PBS–BSA with 8 µg/mL of fl-oxLDL, 20 µM butylated hydroxytoluene, and 100 µM diethylenetriamine penta-acetic acid) in the presence and the absence of test lipids. Again, the plate was incubated for 1 h at ambient temperature. After washing with PBS, the fluorescence value in each well was measured with a plate reader at an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The statistical analysis (one-way analysis of variance and post hoc Dunnett’s test) and the determination of concentration required for 50% inhibition of fl-oxLDL binding (the IC50 concentration) were performed with the aid of a commercially available software (Prism® version 5.0f, GraphPad, San Diego, CA). A p-value less than 0.05 was considered to be statistically significant.
In this study, we aimed at identifying other potential structural elements than carboxyl group recognizable by CD36, by comparing the effect on the fl-oxLDL binding to CD36150–168 of oleic acid with those of lipids with one or more oleyl groups, cholesteryl oleate, triolein, 1,2-diolein 1,3-diolein, 1-monoolein, 2-monoolein, oleamide, and oleic aldehyde. These lipids are known to occur in the diets of human (or of other distinct mammals) or in their bodies as digestsCitation6–8) and thus could often encounter CD36. We added the test lipids to the assay mixture at a fixed concentration (1 mM), referring to that oleic acid caused a potent inhibition around the concentration.Citation5) We measured the effect of oleic alcohol (a negative control lipidCitation5)) under the same conditions.
The results of the assay are presented in Fig. . The data for oleic acid and alcohol were similar to those shown previously.Citation5) Either of the mean fluorescence values obtained using the wells incubated with fl-oxLDL and cholesteryl oleate, triolein, 1,2-diolein, 1,3-diolein, 1-monoolein, 2-monoolein, or oleamide was not substantially lower (even higher in a few of these lipids) than that obtained using the wells incubated with the labeled ligand alone. Indeed, statistical analysis revealed no significant differences in either of the two mean values. Based on the results, we conclude that these lipids exhibit no or little inhibitory effects on the fl-oxLDL binding to CD36150–168 and suggest that cholesteryl, glycerol, and amide (or amino) groups do not primarily serve as elements recognizable by CD36.
Fig. 1. Analysis of the inhibition of fl-oxLDL binding to CD36150–168 by lipids with oleyl group(s).
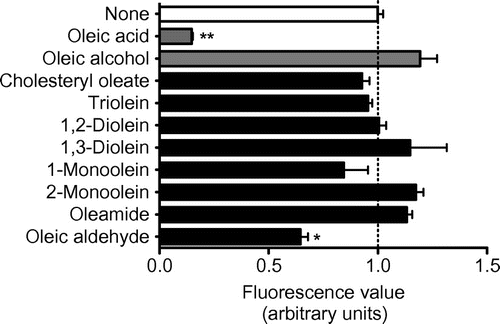
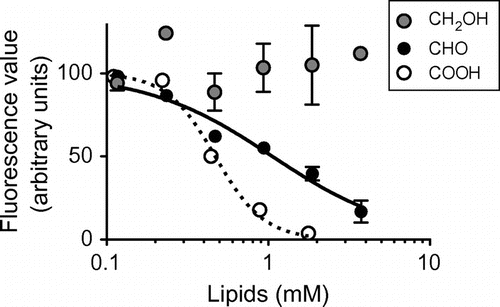
Oleic aldehyde is a long-chain fatty aldehyde that is reportedly present in smoked fish as an odor component.Citation8) The aldehyde is also known to exist in the female gland of Diatraea grandiosella Dyar (a species of moth), although the biological significance in the insect is unclear.Citation9) The mean fluorescence value obtained using the wells incubated with fl-oxLDL and oleic aldehyde was about 65% of and statistically significantly lower than that obtained using the wells incubated with fl-oxLDL alone (Fig. ). We then examined the effects on the binding of fl-oxLDL to CD36150–168 of oleic aldehyde at various concentrations (Fig. ). As references, we presented the data for oleic acid and alcohol (Fig. ). The IC50 concentrations were determined to be 1.1 mM (R2 = 0.93) for oleic aldehyde and 0.47 mM (R2 = 0.99) for oleic acid (not given for oleic alcohol). These results indicated that oleic aldehyde exhibited an inhibitory effect on fl-oxLDL binding to CD36150–168, albeit at lower efficiency than oleic acid. Here, we suggest that a single aldehyde is acceptable as a structural element for recognition by CD36.
Fig. 2. Analysis of inhibition of fl-oxLDL binding to CD36150–168 in the presence of various concentrations of oleic aldehyde.
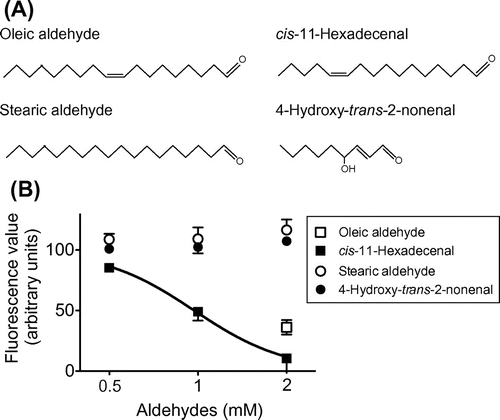
To validate our prediction regarding the effect of an aldehyde group, we finally addressed whether there are additional lipids with a single aldehyde group that inhibit fl-oxLDL binding to CD36150–168. cis-11-Hexadecenal is a long-chain fatty aldehyde that acts as a pheromone in distinct species of insects (Fig. (A)).Citation9) In Drosophila, detection of the aldehyde has been demonstrated to occur via a mechanism that requires sensory neuron membrane protein-1, a homolog of CD36 in the insect.Citation10) As shown in Fig. (B), cis-11-hexadecenal inhibited the binding of fl-oxLDL in a concentration-dependent manner. The IC50 value was determined to be 0.97 mM (R2 = 0.72). This result reinforces our notion about the effect of a single aldehyde. On the other hand, another long-chain fatty aldehyde, stearic aldehyde (known to be used by certain species of insects as a pheromone) (Fig. (A)),Citation11) had no inhibitory effect at any concentrations tested (Fig. (B)). Note that the data for oleic and stearic aldehydes correspond to our previous observations that oleic acid but not stearic acid inhibited the binding of fl-oxLDL to CD36150–168.Citation5) It has been shown that 4-hydroxy-trans-2-nonenal (Fig. (A)), a lipid peroxidation product, displayed no detectable inhibitory effects on the binding of oxLDL to CD36.Citation2) We also had no evidence for inhibition by this hydroxyl fatty aldehyde of fl-oxLDL binding to CD36 mimic (Fig. (B)). Altogether, we propose that an aldehyde group as well as a carboxyl one when linked to distinct hydrocarbon chains could serve as a structural element recognized by CD36. Also, we predict that the acceptability of an aldehyde group by CD36 might associate with the electron-withdrawing ability that depends substantially on the adjacent structure. To define such neighboring structures, we are planning to examine the inhibitory effects of several other lipids with a terminal aldehyde group on the binding of fl-oxLDL to CD36 mimic. At present, we cannot exclude the possibility that higher concentrations of candidate CD36 ligands, including oleic aldehyde, only caused a disruption of fl-oxLDL to inhibit the binding to CD36150–168. Therefore, direct interaction between the candidates and CD36 mimic must also be evaluated.
Fig. 3. Analysis of the inhibition of fl-oxLDL binding to CD36150–168 by the other lipids with a single aldehyde group.
Author contributions
S.T., T.A., and T.F. designed the research plan; S.T. and T.O. performed the fluorescently labeled oxidized low-density lipoprotein binding and competition assay; Y.K. prepared the fluorescently labeled oxidized low-density lipoprotein; T.O. had charge of data processing and computational analysis; S.T. wrote the manuscript with assistance from T.A. and T.O.; T.A. served to procure oleic aldehyde; S.T., T.A., T.O., S.M., K.I., and T.F. analyzed the data; and all authors read and approved the final manuscript.
Funding
This work was supported by the Science and Technology Research Promotion Program for agriculture, forestry, fish- eries and food industry, and JSPS KAKENHI [grant (B) number 25292071] to Tohru Fushiki.
Acknowledgment
We would like to express our sincere gratitude to Mr Yasutaka Ohkubo, R&D Center, T. Hasegawa Co., Ltd., for the preparation of oleic aldehyde.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes
Abbreviations: fl, fluorescently labeled; LCFAs, long-chain fatty acids; oxLDL, oxidized low-density lipoprotein; oxPC, oxidized phosphatidylcholine; PC, phosphatidylcholine.
References
- Horiuchi S, Sakamoto Y, Sakai M. Scavenger receptors for oxidized and glycated proteins. Amino Acids. 2003;25:283–292.10.1007/s00726-003-0029-5
- Podrez EA, Poliakov E, Shen Z, et al. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J. Biol. Chem. 2002;277:38503–38516.10.1074/jbc.M203318200
- Gao D, Ashraf MZ, Kar NS, et al. Structural basis for the recognition of oxidized phospholipids in oxidized low density lipoproteins by class B scavenger receptors CD36 and SR-BI. J. Biol. Chem. 2010;285:4447–4454.10.1074/jbc.M109.082800
- Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 2009;20:72–77.10.1016/j.tem.2008.11.001
- Takai M, Kozai Y, Tsuzuki S, et al. Unsaturated long-chain fatty acids inhibit the binding of oxidized low-density lipoproteins to a model CD36. Biosci. Biotechnol. Biochem. 2014;78:238–244.10.1080/09168451.2014.882750
- Fronimaki P, Spyros A, Christophoridou S, et al. Determination of the diglyceride content in greek virgin olive oils and some commercial olive oils by employing 31 P NMR spectroscopy. J. Agric. Food Chem. 2002;50:2207–2213.10.1021/jf011380q
- Driscoll WJ, Chaturvedi S, Mueller GP. Oleamide synthesizing activity from rat kidney: identification as cytochrome C. J. Biol. Chem. 2007;282:22353–22363.10.1074/jbc.M610070200
- Varlet V, Knockaert C, Prost C, et al. Comparison of odor-active volatile compounds of fresh and smoked salmon. J. Agric. Food Chem. 2006;54:3391–3401.10.1021/jf053001p
- Hedin PA, Davis FM, Dickens JC, et al. Identification of the sex attractant pheromone of the southwestern corn borer Diatraea grandiosella Dyar. J. Chem. Ecol. 1986;12:2051–2063.10.1007/BF01041996
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293.10.1038/nature06328
- Vuts J, Powers SJ, Caulfield JC, et al. Multiple roles of a male-specific compound in the sexual behavior of the dried bean beetle, Acanthoscelides obtectus. J. Chem. Ecol. 2015;41:287–293.10.1007/s10886-015-0560-3
