Abstract
Three putative deuterolysin (EC 3.4.24.29) genes (deuA, deuB, and deuC) were found in the Aspergillus oryzae genome database (http://www.bio.nite.go.jp/dogan/project/view/AO). One of these genes, deuA, was corresponding to NpII gene, previously reported. DeuA and DeuB were overexpressed by recombinant A. oryzae and were purified. The degradation profiles against protein substrates of both enzymes were similar, but DeuB showed wider substrate specificity against peptidyl MCA-substrates compared with DeuA. Enzymatic profiles of DeuB except for thermostability also resembled those of DeuA. DeuB was inactivated by heat treatment above 80° C, different from thermostable DeuA. Transcription analysis in wild type A. oryzae showed only deuB was expressed in liquid culture, and the addition of the proteinous substrate upregulated the transcription. Furthermore, the NaNO3 addition seems to eliminate the effect of proteinous substrate for the transcription of deuB.
Graphical abstract
DeuB was a non-thermostable deuterolysin from A. oryzae. The transcription of the gene was promoted by protein substrates, but the promotion was eliminated by adding inorganic nitrogen source such as NaNO3.
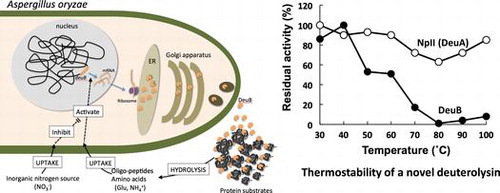
Aspergillus oryzae is one of the well-known fungi used for producing Japanese fermented foods and beverages.Citation1) The micro-organism is been used for more than 1000 years in Japan, and is listed in Generally Regarded as Safe by US Food and Drug Administration.Citation2) The fungus is also famous for producing multiple enzymes and proteins in large amount. Genome sequencing analysis of A. oryzae in 2005 revealed about 12,000 existing genes in the genome.Citation3) About 1% of the genes, 134 genes, were presumed to encode proteolytic enzymes. The number of the proteolytic enzyme genes is larger than that of Aspergillus nidulans and Aspergillus fumigatus whose genome sequences were published concurrently.Citation4,5) The differences were caused by the redundancy of the enzymes categorized into the same group.Citation3) Because of the redundancy, unknown enzymes similar to the known proteolytic enzymes have been found in the genome database of A. oryzae. However, it has not have been clarified why A. oryzae has the redundancy for proteolytic enzymes.
Nakadai et al. showed A. oryzae produces a low molecular mass metalloendopeptidase, NpII.Citation6) The enzyme was distinct from thermolysin-type metalloendopeptidase, NpICitation7) by its molecular mass, substrate specificity, and thermostability. A similar enzyme is found from Aspergillus sojae.Citation8) The enzymes are designated as deuterolysin (EC 3.4.24.39). The molecular mass of the deuterolysins is about 20,000. It is revealed that the enzyme from A. oryzae has a Zn2+-binding and catalytic center motif of -His-Glu-Xaa-Xbb-His-Asp-, and the enzymes from Aspergilli are categorized into aspzincin.Citation9) The enzymes form A. oryzae and A. sojae showed unique thermostability; the enzymes are most unstable at 65–75° C and stable at 100° C.Citation10,11) It was reported that the unique thermostability of the A. oryzae enzyme partially depends on ion bonds between amino acid residues on two α-helixes and a pro residue in a loop structure of the enzyme molecule in comparison with penicillolysin from Penicillium citrinum.Citation12) The enzyme from A. sojae shows high activity against basic nuclear proteins such as histone and protamine and low activity against proteinous substrates generally used in laboratories such as casein, hemoglobin, albumin, and gelatin.Citation13) Doi et al. reported that the deuterolysin from A. oryzae cleaved peptide bonds following dibasic amino acid residues.Citation14)
We found three putative deuterolysin genes in the A. oryzae genome database. One is corresponding to the gene encoding high thermostability enzyme reported by Tatsumi et al.,Citation10) and the other two enzymes have not been reported. We designated the gene encoding the well-known enzyme deuA (AO090010000493), and other newly found genes were designated deuB (AO090001000135) and deuC (AO090701000313). The proteolytic enzymes including the deuterolysin were employed for industry use, especially for food industry, as the fungus and the derived enzymes were assumed to be safe for human consumption. However, deuterolysin seems to cause quality loss of the manufactured foods by the unique thermostability. The deuterolysins are not to be inactive completely even after heat treatment in the manufacturing processes. Therefore, unexpected protein degradation by the remaining deuterolysin can cause changes in the physical properties of the foods. Especially, the remaining proteolytic activity affects fish jelly products from fish surimi with the network structure of myofibrillar proteins by modori phenomenon (the elasticity-lowing).Citation15) Modori phenomenon is closely related with the breakdown of myosin heavy chain, and the breakdown is due to some endogenous proteolytic enzymes.Citation16–18) As the enzymes from A. oryzae are used for producing umami ingredients, addition of the ingredients containing the deuterolysin would accelerate modori phenomenon of fish jelly products. Therefore, an alternative enzyme mixture has been proposed to produce the seasonings for the surimi-based products. As it would be possible to produce the alternative proteolytic enzyme mixture by clarifying the enzymatic profiles and their expression of unknown deuterolysin homologs, we constructed overexpression strains of the three enzymes, and transcriptional analysis of the genes.
Materials and methods
Materials
N-Butoxycarbonyl-L-arginyl-L-valyl-L-argynyl-L-arginyl-4-methylcoumaryl-7-amide (Boc-Arg-Val-Arg-Arg-MCA) and other fluorogenic MCA peptides were purchased from Peptide Institute, Inc. (Ibaraki, Osaka, Japan).
The protein substrates, salmine, BSA, and gelatin, were purchased from Nacalai Tesque Inc. (Kyoto, Japan), and clupeine, histone, ovalbumin, hemoglobin, myoglobin, α-casein, β-lactoglobulin, collagen, elastin, keratin, and zein were purchased from Sigma–Aldrich Co. LLC. (Tokyo, Japan). Casein from bovine milk (acc. to Henmersten) was purchased from MP biomedicals, Inc. (Solon, Ohio, USA). Soybean protein (Fujipro-AL) was kindly gifted by Fuji oil Co. Ltd. (Izumisano, Japan).
Strains and culture conditions
A. oryzae RIB40 (ATCC-42149)Citation19) was used as the gene donor for analysis of transcription of deuterolysin genes. A. oryzae niaD300 (niaD-)Citation20) was used to construct overexpression strains of deuterolysin. Escherichia coli DH5α (supE44, ΔlacU169 (φ80lacZΔM15), hsdR17, recA1, endA1, gyrA96, thi-1, relA1) was used for DNA manipulation.
The various media used for transcriptional analysis are Czapeck-Dox (CDN) medium,Citation21) CDN medium with 0.3% skimmed milk (CDNS), CDN medium with 0.6% skimmed milk instead of 0.6% NaNO3 (CDS), and CDN medium with 0.6% gelatin instead of 0.6% NaNO3 (CDG). YPD medium (1% yeast extract, 2% polypeptone, and 3% glucose) was used for preparing genomic DNA and maintaining the strains. YPM medium (1% yeast extract, 2% polypeptone, and 3% maltose) was used for overexpression of the recombinant enzyme.
Plasmid construction
All the following basic molecular biology procedures were carried out as described by Sambrook et al.Citation22) The DNA fragment of deuterolysin genes, deuA, deuB and deuC, was amplified by PCR with A. oryzae RIB40 genome DNA as a template employing primer sets of deuA-F and deuA-R, deuB-F and deuB-R, and deuC-F and deuC-R, respectively. The primer sequences are shown in Table . The amplified DNA fragments were digested with NdeI and XbaI and ligated with a plasmid, pNGA142Citation23) digested with NdeI and XbaI. The nucleotide sequence of the inserted fragment was confirmed using ABI 310 Genetic Analyzer (Applied Biosystems Inc, Foster City, CA, USA). The resulting DeuA, DeuB, and DeuC overexpression vectors were named pNGdeuA, pNGdeuB, and pNGdeuC, respectively.
Table 1. The primers used in this study.
Construction of overexpressing strains
A. oryzae niaD300 was used as a host strain for constructing deuterolysin overexpressing strains. The overexpression vector, pNGdeuA, pNGdeuB, or pNGdeuC, was introduced into the host strain by protoplast polyethlenglycol method.Citation24) The colonies grown on CDN plate were picked up and then the recombination was confirmed by PCR using primer pairs of deuAC, deuBC or deuCC, and niaDC (Table ) with the candidate genome DNA as a template.
Proteolytic Activity Assay
Proteolytic activities with salmon protamine sulfate (salmine) were assayed at pH 7.0 and 30° C. Fifty microliters of sample and 400 μL of 100 mM sodium phosphate buffer, pH 7.0, were mixed and pre-incubated for 5 min, and then 150 μL of 2% (w/v) salmon protamine sulfate (previously denatured at 100° C for 30 min in 100 mM sodium phosphate buffer, pH 7.0) was added and incubated for various periods. Six-hundred microliters of 12.5% (w/v) trichloroacetic acid solution containing 20% (w/v) NaCl was added to terminate the enzyme reaction, and then the mixture was filtered with Advantec No. 2 filter paper (Advantec Toyo Kaisha, Ltd., Tokyo, Japan). Two-hundred micro-liters of the filtrate and 3.0 mL of 0.5 M sodium citrate buffer (pH 5.0) were mixed, and 1 mL of freshly prepared ninhydrin reagentCitation25) was also added. The linearity of the assay was checked, and the amount of amino acid produced in the reaction mixture was determined. One katal is defined as the amount of enzyme yielding the color equivalent of 1 mol of tyrosine in 1 s with ninhydrin reagent using protein substrates at pH 7.0 and 30° C according to previous work.Citation25)
SDS-polyacryl amide gel electrophoresis
SDS-PAGE was performed with 15% acrylamide gel as described previously.Citation26)
Purification and analysis of overexpressed enzymes
The conidia of each overexpression strain were inoculated in 200 mL of YPM liquid medium, adjusted the concentration to 5 x 105 conidia/mL, and cultivated aerobically at 30° C for 36 h. The culture broths were collected by filtration with Miracloth™ (Merk Japan Ltd., Tokyo, Japan). For purifying DeuA, (NH4)2SO4 was added to the culture broth slowly up to 70% saturation and left overnight. Then, the solution was centrifuged at 4° C, 12,000 g for 30 min. The supernatant was applied to a phenyl-Sepharose CL-4B column chromatography (ø30 x 130 mm) equilibrated with 50 mM acetate buffer, pH 6.0, containing 70% saturated (NH4)2SO4. The column was washed with the same buffer extensively, and the enzyme was eluted by a 70–0%-saturated (NH4)2SO4 gradient. The active fraction was collected and dialyzed against 50 mM acetate buffer, pH 6.0. For purifying DeuB, the culture broth was dialyzed against 10 mM acetate buffer, pH 5.0, and then the dialysate was applied to the CM-cellulofine C-500 column chromatography (φ35 x 85 mm) equilibrated with the same buffer. The column was washed with the same buffer extensively. The enzyme was eluted with 0–0.4 M NaCl linear gradient. The active fraction was collected and dialyzed against the same buffer.
N-terminal amino acid sequence
The purified enzymes subjected to SDS-PAGE were transferred onto a PVDF membrane using a Trans-Blot SD semi-dry transfer cell (Bio-Rad Laboratories, Hercules, CA, USA). The proteins were stained with Coomasie Brilliant Blue R-250 and excised from the blot.Citation27) A piece of the blots was analyzed using an Applied Biosystems® 473A protein sequencer with a 610A data analysis system (Applied Biosystems Inc., Foster City, CA, USA).
Optimum pH and pH stability
The effect of pH on enzymatic activities was measured in the pH range of 3.0–11.0. The pH stability of the enzymes was studied in the pH rage 3.0–12.0. The enzymes were incubated in 100 mM Briton-Robinson broad range buffer at each pH and 4° C for 12 h and then the enzymatic activities for salmine were measured at 30° C as described above.
Optimum temperature and thermostability
The effect of temperature on enzymatic activities was measured in the temperature range of 30–100° C for 5 min. Thermostability of the activities was measured after incubation in the same temperature range for 10 min and then iced quickly. The enzyme activity was measured as described above.
Substrate specificity
Proteolytic activity of two deuterolysins, DeuA and DeuB, against protein substrates was examined according to the proteolytic activity assay method using not only salmine but also herring protamine (clupeine), histone, BSA, ovalbumin, hemoglobin, myoglobin, α-casein, β-lactoglobulin, casein, gelatin, collagen, elastin, fibrin, keratin, zein, and soy protein as substrates.
Activities against fluorogenic MCA-substrates were measured by a modified method described before.Citation28) Five microliters of the each MCA-substrate dissolved in dimetylsulfoxide was added to 1.3 mL of 100 mM phosphate buffer, pH 7.0, pre-incubated at 30° C. Then, 95 μL of the diluted enzyme solution was added to the substrate solution. Increment of fluorescence from released 7-amino-4-methyl coumarine was measured with Fluorescence Spectrophotometer F-3000 (Hitachi Ltd., Tokyo, Japan) with an excitation of 340 nm and an emission of 440 nm.
Transcriptional analysis of deuterolysin genes
All molecular manipulations given below were performed according to the manufacturers’ instructions. Total RNA from hyphae cultured under various conditions was extracted using Sepasol-RNA I SuperG (Nacalai Tesque Inc., Kyoto, Japan). The genomic DNA was eliminated using RNase-free DNase I (Takara Bio Inc., Kusatsu, Japan), while messenger RNA (mRNA) was purified using the Oligotex-dT30 Super mRNA purification kit (Takara Bio Inc., Kusatsu, Japan). Complementary DNA (cDNA) was synthesized using the TaKaRa RNA PCR Kit (AMV) Ver3.0 (Takara Bio Inc., Kusatsu, Japan) with oligo-dT primers.
Semi-transcription quantity was estimated using PCR with the primer pairs: T-deuA-F and T-deuA-R, T-deuB-F and T-deuB-R, and T-deuC-F and T-deuC-R, for deuA, deuB, and deuC cDNA, respectively. The semi-quantities PCR under each culture condition were normalized with the quantity of β-actin transcription, estimated with PCR amplification with the primer pair of actin-F and actin-R, and each cDNA.
Results
Three deuterolysin genes, AO090010000493, AO090001000135, and AO090701000313, were found in A. oryzae genome database (http://www.bio.nite.go.jp/dogan/project/view/AO) and were designated as deuA, deuB, and deuC, respectively. The presumed coding regions of deuA, deuB, and deuC were 1175, 1165, and 1077 bp, respectively. We cloned the genes and constructed overexpressing strains using A. oryzae niaD300 as a host. The integration of the overexpression plasmid for deuA, deuB, and deuC in the niaD locus of the host genome was confirmed by PCR (Fig. S1) and the amplified DNA fragments of about 5.0 kbp were observed in every strain. One of the confirmed transformants was used in the following experiments. The resulting overexpression strains were designated as A. oryzae deuAOE, deuBOE, and deuCOE, and cultured in YPM medium aerobically. As the result of it, proteins, which could not be seen in the culture broth of the control strain, were observed at about 22 k of molecular mass in those of deuAOE and deuBOE strains (Fig. ). However, the protein corresponding to deuCOE was not observed in the culture broth. We purified DeuA and DeuB as described above, and the purified enzymes were applied to SDS-PAGE. The results are shown in Fig. . As shown in lane 1 of (A) and (B), DeuA and DeuB are shown as a single band of 23 k and 21 k of molecular masses, respectively. The N-terminal amino acid sequence of the purified DeuA, 1Thr-Glu-Val-Thr-5Asp- (Fig. S2), was identical with a previous report.Citation10) Meanwhile, N-terminal sequence of DeuB was 1Thr-Lys-Val-Thr-5Ser- (Fig. S3). As the results of it, DeuB consisted of 20 amino acid residues of signal peptide projected by Signal IP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/), 156 residues of propeptide, and 177 residues of mature enzyme region. It was similar to DeuA, but only the length of signal peptide was different.
Fig. 1. SDS-PAGE of the culture broth from each overexpression strain for deuterolysin homologs.
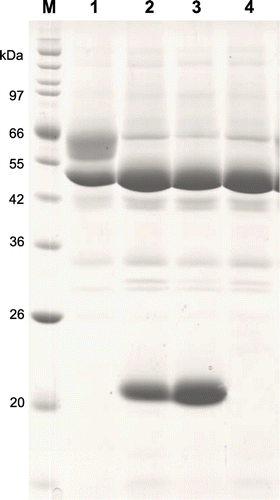
Fig. 2. SDS-PAGE of purified DeuA and DeuB from the overexpression strains.
The substrate specificity of the enzymes was determined. As shown in Table , DeuA and DeuB showed the highest specific activity against clupeine; 100 and 73 mkat/kg protein, respectively, followed by salmine and histone. Other than these nucleoproteins, both enzymes could digest α-casein, ß-lactoglobulin, ovoalbumin, fibrin, and casein even at lower levels compared with nucleoproteins. Though DeuA could not digest elastin and collagen, DeuB could not digest only collagen. Furthermore, we examined the substrate specificity using fluorogenic peptide MCA-substrates. As shown in Table , the most favorable substrate for DeuA and DeuB was Boc-Arg-Val-Arg-Arg-MCA. The second-best substrate for DeuB was Z-Val-Lys-Met-MCA, but DeuA could not hydrolysis it. Like this substrate, DeuA could not digest Z-Leu-Arg-MCA, Z-Val-Val-Arg-MCA and Suc-Gly-Pro-MCA, which were digested by DeuB. Furthermore, DeuB could digest Glt-Ala-Ala-Phe-MCA, Suc-Leu-Val-Tyr-MCA, and Boc-Val-Leu-Lys-MCA relatively well, but DeuA showed lower specific activities. These results showed these enzymes had a similar digestion property against the protein substrates, but DeuB had a wider substrate specificity compared with DeuA.
Table 2. Substrate specificities of DeuA and DeuB toward protein substrates.
Table 3. Substrate specificities of DeuA and DeuB against fluorogenic peptidyl MCA-substrates.
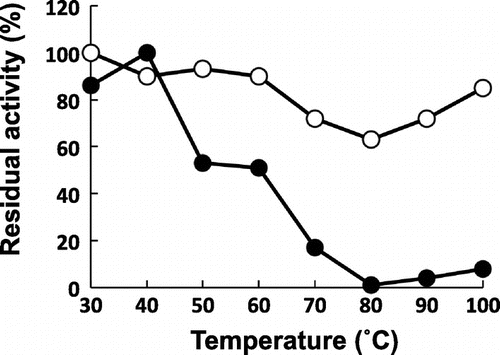
Optimum temperatures of the DeuA and DeuB were 50–60 and 50–70° C, respectively, and optimum pH for both enzymes was 6–10. Both enzymes showed high tolerance against pH change; stable pH of both enzymes was 3–11. Thermostability of the enzymes was different (Fig. ). DeuA showed high thermostability as previously reported. On the other hand, DeuB lost its activity at 80° C, but a small amount of activity was detected over 80° C.
Fig. 3. Themostability of DeuA and DeuB.
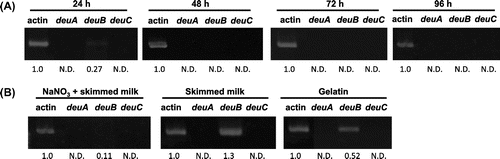
The transcription of three genes in the wild type A. oryzae strain was examined in the liquid culture with various nitrogen sources, as shown in Fig. . The transcription of deuA and deuC was not observed in any condition we examined. On the other hand, the transcription of deuB was observed at an early stage of CDN liquid culture (Fig. (A), 24 h) and the prolonged cultivation reduced the transcription of deuB. The proteinous nitrogen sources such as skimmed milk and gelatin instead of NaNO3 upregulated the transcription level of deuB, respectively (Fig. (B), skimmed milk and gelatin). However, the addition of NaNO3 inhibited the upregulation of deuB transcription (Fig. (B), NaNO3 + skimmed milk).
Fig. 4. Semi-quantitative transcription analysis of deuA, deuB, and deuC: A. oryzae RIB40 was cultivated in CDN medium for 24, 48, 72, and 96 h (A) and in CDNS, CDS, and CDG media for 24 h (B).
Discussion
We have been attempted to clarify the reason of redundancy on the proteolytic enzyme genes in A. oryzae through substrate specificity of the proteolytic enzymes and their expression patterns. In this study, we found three deuterolysin homolog genes in the genome database of A. oryzae and the genes were designated deuA (AO090010000493), deuB (AO090001000135), and deuC (AO090701000313). The gene deuA corresponds to the NpII gene previously reported by Tatsumi et al.Citation10) We constructed the overexpression strain for each enzyme, and the expression of DeuA and DeuB was confirmed. However, the DeuC was not detected in the culture broth of the overexpression strain.
The purified DeuB showed a similar digestion profile to that of DeuA against the protein substrates. However, DeuB showed a wider substrate specificity against fluorogenic peptidyl MCA-substrates compared with DeuA, as DeuB could digest all the peptidyl MCAs. Doi et al. reported that DeuA may recognize dibasic amino acids at P2-P1 of the substrate and P3 is also important in the recognition of peptidyl MCA-substrate,Citation14) but the tendency by DeuA is not observed for the protein substrate such as a histone. DeuB could digest all of the peptidyl MCA-substrates that we examined. As the enzyme could accept Glu, Leu, and Met residues at P1 position, DeuB might recognize the aliphatic carbon(s) of side chain of amino acid residue at P1 position. Optimum pH and stable pH of DeuB were similar to those of DeuA, but thermostability of DeuB was different from that of DeuA. DeuB lost its enzymatic activity above 80° C. It is a very important profile of the enzyme for the food industry. It indicates DeuB in food products can be inactivated by heat treatment below 100° C. Modori phenomenon in fish jelly products caused by DeuA should be prevented using DeuB instead of DeuA for producing seasonings to add to the manufactured food. Doi et al. reported that thermostability of DeuA partially depends on an ionic bonding between the Glu30 and Arg61 residues on separate α-helixes and the Pro167 residue in a loop structure by the structure comparison between DeuA and penicillolysin from Penicillium citrinum.Citation12) In DeuB, residues of Gln30, Asn61, and Val167 were corresponding to Glu30, Asp61, and Pro167 of DeuA. These differences seemed to be the reason of lower thermostability.
Furthermore, our transcription analysis indicated that deuA was not transcribed in the liquid culture, even though the protein substrates existed. Meanwhile, deuB was translated at an early stage in CDN liquid culture and the transcription was upregulated by the proteinous substrates. Interestingly, the upregulated transcription by the protein substrates was eliminated by the addition of NaNO3. A transcription factor, AreA, activates the uptake of inorganic nitrogen source, NO3- or NO2-, and the uptake is repressed by Glu and NH4+ in A. nidulans.Citation29) It should mean the Glu released from proteins degraded by proteolytic enzymes represses the uptake of NO3- or NO2- and A. nidulans gives priority to utilize organic nitrogen source than inorganic nitrogen source. On the basis of the facts, A. oryzae should also prefer organic nitrogen substrates to inorganic ones, but our results seemed to indicate NaNO3 repressed the production of DeuB. DeuB might not be necessary in that situation. The same tendency was observed in the transcription of dppB, one of the three dipeptidyl peptidase genes in A. oryzae.Citation30)
It would be possible to produce the enzyme preparation containing DeuB instead of DeuA by the cultivation of A. oryzae in liquid medium with proteinous substrates without inorganic nitrogen source. Tatusmi et al. obtained the mRNA of deuA from the culture with wheat bran, and they could not found DeuB from the culture.Citation10) It was thought that A. oryzae might express DeuA and DeuB according to the culture conditions of liquid or solid. The same occurrence is shown in the case of glucoamylases, GlaA and GlaB, of A. oryzae.Citation31,32) GlaA was expressed in the liquid culture and GlaB was observed in the solid culture. In the expression database of CAoGD (http://nribf21.nrib.go.jp/CFGD/), high transcription of deuA was observed in solid culture (rice koji and solid wheat bran culture) at an early growth stage until 24 h by DNA microarray analysis. In the wheat bran liquid culture, about 1/60 of the transcription in the solid culture was observed at 24 h and then reduced. It was also reported that the transcription of deuA was not found by the expression sequence tag analysis for the conidia and the germinated conidia.Citation21) On the other hand, the transcription level of deuB in rice koji was less than 1/250 of the deuA transcription, but high transcription was observed in the solid wheat bran culture. It seems that deuA would transcribe in the solid culture like glaB. The transcription of deuB might be induced by proteinous substrates in both solid and liquid cultures, as wheat bran is composed of 16–17% proteins (dry basis).Citation33) The fact suggests that enzyme preparation, which does not cause the modori phenomenon of fish jelly products, is possible by the liquid culture containing proteinous substrates without inorganic nitrogen sources.
Authors contribution
Y. Y. conceived the project, designed the experiments, and wrote the manuscript. M. H., T. K., and D. S. performed the experiments and wrote the manuscript. M. T, K. K., H. A., H. I., and K. A. were involved in study conception and interpretation of data.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental materials
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2016.1166933.
TBBB_1166933_Supplementary_data.pdf
Download PDF (255.8 KB)References
- Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of koji mold and exploration of its future. DNA Res. 2008;15:173–183.10.1093/dnares/dsn020
- Taylor MJ, Richardson T. Applications of microbial enzymes in food systems and in biotechnology. Adv. Appl. Microbiol. 1979;25:7–35.10.1016/S0065-2164(08)70144-8
- Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161.10.1038/nature04300
- Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115.10.1038/nature04341
- Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156.10.1038/nature04332
- Nakadai T, Nasuno S, Iguchi N. Purification and properties of neutral proteinase II from Aspergillus oryzae. Agric. Biol. Chem. 1973;37:2703–2708.10.1271/bbb1961.37.2703
- Nakadai T, Nasuno S, Iguchi N. Purification and properties of neutral proteinase I from Aspergillus oryzae. Agric. Biol. Chem. 1973;37:2695–2701.10.1271/bbb1961.37.2695
- Sekine H. Neutral proteinases I and II of Aspergillus sojae. Agric. Biol. Chem. 1972;36:198–206.10.1271/bbb1961.36.198
- Fushimi N, Ee CE, Nakajima T, et al. Aspzincin, a family of metalloendopeptidases with a new zinc-binding motif: identification of new zinc-binding sites (His128, His132, and Asp164) and three catalytically crucial residues (Glu129, Asp143, and Tyr106) of deuterolysin fromaspergillus oryzae by site-directed mutagenesis. J. Biol. Chem. 1999;274:24195–24201.10.1074/jbc.274.34.24195
- Tatsumi H, Murakami S, Tsuji RF, et al. Cloning and expression in yeast of a cDNA clone encoding Aspergillus oryzae neutral protease II, a unique metalloprotease. Mol. Gen. Genet. 1991;228:97–103.
- Sekine H. Neutral proteinases I and II of Aspergillus sojae. Some enzymatic properties. Agric. Biol. Chem. 1972;36:207–216.10.1271/bbb1961.36.207
- Doi Y, Akiyama H, Yamada Y, et al. Thermal stabilization of penicillolysin, a thermolabile 19 kDa Zn2+-protease, obtained by site-directed mutagenesis. Protein Eng. Des. Sel. 2004;17:261-266.
- Sekine H. Neutral proteinase II of Aspergillus sojae: an enzyme specifically active on protamine and histone. Agric. Biol. Chem. 1973;37:1765–1767.10.1271/bbb1961.37.1765
- Doi Y, Lee BR, Ikeguchi M, et al. Substrate specificities of deuterolysin from Aspergillus oryzae and electron paramagnetic resonance measurement of cobalt-substituted deuterolysin. Biosci. Biotechnol. Biochem. 2003;67:264-270
- Toyohara H, Shimizu Y. Relation between the modori phenomenon and myosin heaby chain breakdown in Threadfin-bream gel. Agric. Biol. Chem. 1998;52:255–257.
- Kinoshita M, Toyohara H, Shimizu Y. Diverse distribution of four distinct types of Modori (gel degradation)-inducing preoteinases among fish species. Nippon Suisan Gakk. 1990;56:1485–1492.10.2331/suisan.56.1485
- Hu Y, Ji R, Jiang H, et al. Participation of cathepsin L in modori phenomenon in carp (Cyprinus carpio) surimi gel. Food Chem. 2012;134:2014–2020.10.1016/j.foodchem.2012.04.060
- Yoshida A, Ohta M, Kuwahara K, et al. Purification and characterization of cathepsin B from the muscle of horse mackerel Trachurus japonicus. Mar. Drugs. 2015;13:6550–6565.10.3390/md13116550
- Machida M. Progress of Aspergillus oryzae genomics. Adv. Appl. Microbiol. 2002;51:81–106.10.1016/S0065-2164(02)51002-9
- Minetoki T, Nunokawa Y, Gomi K, et al. Deletion analysis of promoter elements of the Aspergillus oryzae agdA gene encoding α-glucosidase. Curr. Genet. 1996;30:432–438.10.1007/s002940050153
- Tsujii M, Okuda S, Ishi K, et al. A long natural-antisense RNA is accumulated in the conidia of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2016;80:386–398.10.1080/09168451.2015.1101333
- Sambrook J, Fritsh EF, Maniatis T. Moleccular cloning: a laboratory manual. 2nd ed. Spring Hoarbor (NY): Cold Spring Harbor Laboratory Press; 1989.
- Minetoki T, Tsuboi H, Koda A, et al. Development of high expression system with the improved promoter using the cis-acting elements in Aspergillus species. J. Biol. Macromol. 2003;3:89–96.
- Mizutani O, Nojima A, Yamamoto M, et al. Disordered cell integrity signaling caused by disruption of the kexb gene in Aspergillus oryzae. Eukaryot. Cell. 2004;3:1036–1048.10.1128/EC.3.4.1036-1048.2004
- Yamaguchi M, Hanzawa S, Hirano K, et al. Specificity and molecular properties of penicillolysin, a metalloproteinase from Penicillium citrinum. Phytochemistry. 1993;33:1317–1321.10.1016/0031-9422(93)85082-3
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685.10.1038/227680a0
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038.
- Yamagata Y, Ichishima E. A new alkaline proteinase with pI 2.8 from alkalophilic Bacillus sp. Curr. Microbiol. 1989;19:259-264.
- Tudzynski B. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol. 2014;5:656.
- Maeda H, Sakai D, Kobayashi T, et al. Three extracellular dipeptidyl peptidases found in Aspergillus oryzae show varying substrate specificities. Appl. Microbiol. Biotechnol. 2016. [Epub ahead of print].
- Ishida H, Hata Y, Ichikawa E, et al. Regulation of the glucoamylase-encoding gene (glaB), expressed in solid-state culture (koji) of Aspergillus oryzae. J. Ferment. Bioengneer. 1998;86:301–307.10.1016/S0922-338X(98)80134-7
- Ishida H, Hata Y, Kawato A, et al. Identification of functional elements that regulate the glucoamylase-encoding gene (glaB ) expressed in solid-state culture of Aspergillus oryzae. Curr. Genet. 2000;37:373–379.10.1007/s002940000118
- D’Appolonia BL. Use of nonfluor fraction of wheat. Cereal Food. World. 1979;24:326–331.
