Abstract
Promoter shutoff is a general method for analyzing essential genes, but in the fungus Aspergillus oryzae, no tightly repressed promoters have been reported. To overcome the current limitations of conditional promoters, we examined sorbitol- and galactose-responsive genes using microarrays to identify regulatable genes with only minor physiological and genetic effects. We identified two sorbitol-induced genes (designated as sorA and sorB), cloned their promoters, and built a regulated egfp and brlA expression system. Growth medium-dependent enhanced green fluorescence protein (EGFP) fluorescence and conidiation were confirmed for egfp and brlA under the control of their respective promoters. We also used this shutoff system to regulate the essential rhoA, which demonstrated the expected growth inhibition under repressed growth conditions. Our new sorbitol promoter shutoff system developed can serve as a valuable new tool for essential gene analyses of filamentous fungi.
Graphical abstract
A sorbitol-responsive promoter shutoff system was evaluated for egfp and essential rhoA expression controlled with PsorA and PsorB in A. oryzae.
As a “national organism” of Japan, the fungus Aspergillus oryzae is one of the most important microorganisms used in the Japanese fermentation industry to create sake, miso, and soy source products, thus contributing to the rich culture of Japanese food.Citation1) As a result of its high protein production and safety, A. oryzae has recently attracted wide industry attention as a host organism for heterologous protein production. Given the importance of this fungus to the fermentation industry, several studies have evaluated A. oryzae gene expression with the goal of developing inducible promoters from Aspergillus species.
Functional gene expression analyses have been performed by substituting an original promotor with an inducible promoter and then inducing or repressing the target gene under specific conditions.Citation2) At present, there are several inducible promoters such as the alcohol-inducible alcA promoter (PalcA),Citation3) the starch- and maltose-inducible amyB promoter (PamyB),Citation4) the thiamine-repressible thiA promoter (PthiA),Citation5) and others.Citation6,Citation7,Citation8) The PalcA system does not function well within A. oryzae and is not commonly used; however, one study has reported it as an inducible promoter.Citation9) The PthiA system is tightly regulated with a riboswitch but becomes inactive under alkaline growth conditions.Citation5) The other existing promoter systems do not completely repress gene expression, and switching between conditions for induction and repression often has pleiotropic effects on intracellular metabolism.Citation10) It is thus necessary to develop a promoter shutoff system with an inducible promoter that is strictly repressed under shutoff conditions that have minimal effects on the regulation of other genes. The availability of a strict promoter shutoff regulation system would enable essential gene analyses.
In this study, we focused on the D-galactose and D-sorbitol metabolism pathways, which feed into the D-glucose glycolytic pathway through the Leloir pathwayCitation11) and the oxide-reductive pathway,Citation12) respectively. We used microarrays to examine the A. oryzae RIB40 gene expression under multiple mycelia culture conditions. The gene expression was evaluated with D-glucose as a repressed culture condition and compared to the expression in D-galactose and D-sorbitol as an induction culture condition. Our results indicated lesser whole gene expression than expected; however, we identified two genes (designated as sorA and sorB) that were highly expressed under the induction culture conditions but marginally expressed or not expressed under the repressed culture conditions.
To evaluate the ability of the two gene promoters to regulate the gene expression, we developed enhanced green fluorescence protein (EGFP) expression systems regulated by the sorA (PsorA) and sorB (PsorB) promoters. In addition, we placed brlA, a key conidiation transcription factor-encoded gene, under the regulation of PsorA-S, PsorA-L, and PsorB to evaluate the promoter shutoff ability in A. oryzae. We also created an inducible construct of PsorA-S-rhoA, an essential regulator of beta-1,3-glucan synthase, to apply this new promoter shutoff system for the functional analysis of essential genes. This promoter shutoff system should prove useful for additional analyses of functionally essential genes.
Materials and methods
Strains and cultivation
A. oryzae strains RIB40 and NSR-ΔLD2 were used for the microarray analysis and as host cells for gene modification. Czapeck-Dox medium was used for the general mycelia cultivation. MM medium [1 g glucose, 3 g glutamate, 2 g NH4Cl, 1 g (NH4)2SO4, 0.5 g KCl, 1 g KH2PO4, 0.5 g MgSO47H2O, 0.02 g FeSO4, and 1.5 g L-methionine in 1 L; pH 6.5] was used for the pre-culture and repressive culture. GM (MM + 1% galactose without glucose) and SM (MM + 1% sorbitol without glucose) media were used for the gene expression analyses as well as the induction and repression experiments. The strains used in this study are listed in Table .
Table 1. Strains used in this study.
RNA preparation for gene expression analysis
A. oryzae RIB40 were incubated in 100 mL of MM medium at 100 rpm for 24 h at 30 °C. Subsequently, mycelia were recovered by Miracloth filtration material (Calbiochem, Darmstadt, Germany) and transferred in 100 mL of each of the induction media used (MM, GM, and SM). With each medium, cultures were performed at 100 rpm for 1 h at 30 °C. The mycelia were harvested by Miracloth filtration, washed twice with ice-cold water, and dried with paper towels. The mycelia were then frozen in liquid N2 and finely ground. Total RNA was extracted from the mycelia using Isogen (Wako, Osaka, Japan) according to the manufacturer’s instructions. General RNA handling was in accordance with “Molecular Cloning.”Citation13)
Microarray analysis
We performed the GeneChip microarray analysis using an established procedure as described.Citation14) We used the A. oryzae GeneChip microarray [AoDNAChip; NCBI Gene Expression Omnibus (GEO) platform GPL16184] designed by Affymetrix (Santa Clara, CA) for the transcriptome analysis of 13,765 ORF probes included in the microarray set.
We used an RNeasy Mini kit (Qiagen, Hilden, Germany) to purify the total RNA for microarray analysis. The RNA quality was determined using a Bioanalyzer 2100 system (Agilent Technology, Santa Clara, CA), and the RNA quantity was determined using an Ultrospec 3300 Pro spectrophotometer (Amersham Pharmacia Biotech, Buckinghamshire, UK). We prepared fragmented biotin-labeled cRNA using a GeneChip one-cycle target labeling and control reagent kit (Affymetrix) according to the manufacturer’s instructions. The fragmented cRNA was hybridized to an AoDNAChip. The GeneChip was then washed, stained, and scanned using a GeneChip FS-450 fluidics station (fluidics protocol FS450_001) and a GeneChip 3000 scanner.
Scanned probe array images were converted into CEL files and normalized using the program GCOS v.1.4 (Affymetrix). Signal intensity and detection p-value calculations were also performed using GCOS v.1.4. The trimmed mean signal of the array was scaled to a target signal of 500 using the all probe sets scaling option. The detection call was used for the detection of a particular transcript, with a detection p-value < 0.04 considered present (P), 0.04 ≤ p<0.06 considered marginal (M), and a p-value of ≥ 0.06 considered absent (A).
These calculation data were also exported as CHP files. For the microarray data analysis, the CHP files were imported into GeneSpring v.7.3 (Agilent Technologies), and gene expression data were normalized per chip to the 50th percentile. We identified genes with statistically significant changes in transcript abundance using a cutoff value of twofold and a Welch’s t-test value < 5%. Two biological replicates were used in the microarray analysis. We submitted the microarray data to the GEO (GEL76534).
Confirmation of the transcriptional start site for each gene
We performed the 5′-rapid amplification of cDNA ends (5′-RACE) using the SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions and the primer set of the 10 × Universal Primer A Mix (Fw. in the kit) and SorA or SorB (Rev.) (Table ). We sub-cloned several amplified fragments into pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA) and sequenced using M13 primer sets.
Table 2. Primers used in this study.
Construction of sorA and sorB promoters::EGFP expression strains
PsorA-S (AO090020000639), PsorA-L, and PsorB (AO090020000635) were amplified with the primer sets 029-697LP and 026-1RP, 026-1363LP and 026-1RP, and 023-611LP and 023-1RP, respectively (Table ). Each polymerase chain reaction (PCR)-amplified fragment (PsorA-S, PsorA-L, and PsorB) was sub-cloned into pCR-BluntII-TOPO (Invitrogen) according to the manufacturer’s instructions. Promoter regions were digested by Kpn I and ligated into pUSACitation15) at the Kpn I site. We amplified the region from each promoter to the TamyB site using Fusion-USC-26S-F (for PsorA-S), Fusion-USC-26L-F (for PsorA-L), Fusion-USC-23-F (for PsorB), and Fusion-USC-amyT-R primer combinations with KOD-Plus-Neo (Toyobo, Osaka, Japan).
We cloned the amplified fragments into pUSCCitation16) digested with Sma I using the In-Fusion HD cloning kit (Clontech) according to the manufacturer’s instructions to construct pUSC-PsorA-S-TamyB, pUSC-PsorA-L-TamyB, and pUSC-PsorB-TamyB. We amplified egfp fragments with Fus-promoter-TamyB-F and Fus-promoter-TamyB-R. pUSC-PsorA-L-TamyB was digested with Sma I, and we ligated the amplified fragment using the Infusion HD cloning kit and labeled it as pUSC-PsorA-L-T-EGFP. The other promoters were similarly constructed and labeled pUSC-PsorA-S-T-EGFP and pUSC-PsorB-T-EGFP. Each constructed plasmid was digested with Cla I or Aor51HI, transformed to NSR-ΔLD2Citation17) by the protoplast-PEG method,Citation18) and cultured in MM medium lacking methionine. The transformants were then subcultured for two passages and verified by PCR analysis.
Construction of the brlA shutoff strain
We constructed a brlA promoter shutoff cassette using the fusion PCR method.Citation19) The L-arm and R-arm of brlA (AO090005001041) were amplified with the use of BrlA-sC-F and BrlA-sC-R, PsorA-S-BrlA-F or PsorB-BrlA-F and Pro-BrlA-R, respectively (Table ). The sC marker and each promoter region were amplified with sC-Pro-F and PsorA-S-sC-PsorA-R or PsorB-sC-PsorB-R primer sets as a template for pUSC-PsorA-S-T-EGFP and pUSC-PsorB-T-EGFP, respectively (Table ). The three amplified fragments were mixed, and a fusion PCR was performed using Nested-PsorA-S-BrlA-F2 and R2, Nested-PsorB-BrlA-F1, and R1. The fused brlA shutoff cassettes were then transformed into the NSR-ΔLD2 strain. The transformants were then subcultured for two passages and verified by PCR analysis.
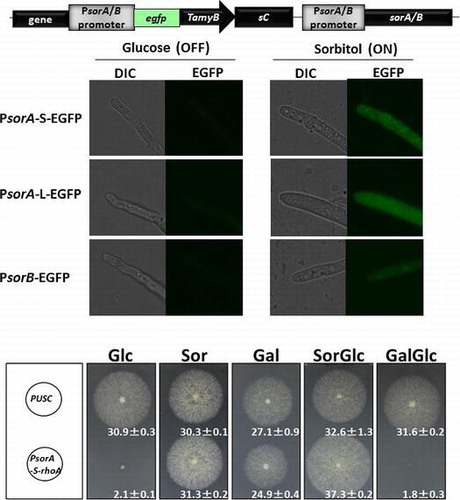
From each strain 1 × 105 spores (in 0.5 μL) were point-inoculated on MM and SM agar plates and incubated for 6 days at 30 °C. Images of each plate were captured using a SteREO Lumar.V12 stereoscope (Zeiss, Jena, Germany). We extracted the conidia present on each plate using 0.1% Tween in a 0.9% NaCl solution, and we counted them using a TC10 automated cell counter (Bio-Rad, Hercules, CA). The colony diameter and the number of conidia formed per cm2 colony were determined.
Construction of the rhoA shutoff strain
We constructed a rhoA promoter shutoff cassette by the fusion PCR method. The L-arm and R-arm of rhoA (AO090701000044) in A. oryzae RIB40 were amplified using AorhoA-sC-F and AorhoA-sC-R and PsorA-S-AorhoA-f and Pro-AorhoA-R, respectively (Table ). The sC marker and the PsorA moiety were amplified using sC-Pro-F and PsorA-S-sC-PsorA-R as templates for pUSC-PsorA-S-T-EGFP. The three amplified fragments were mixed, and a fusion PCR was performed using Nested-PsorA-S-AorhoA-F1 and R1. Fused rhoA shutoff cassettes were transformed into the NSR-ΔLD2 strain. The transformants were subcultured for two passages and verified by PCR analysis. From each strain, 1 × 104 spores (in 1 μL) were point-inoculated on MM, GM, SM, SorGlc (MM with 1% sorbitol), and GalGlc (MM with 1% galactose) agar plates and incubated for 3 days at 30 °C.
Fluorescence microscopy
Conidia from EGFP expression strains (1.0 × 108 cell) were inoculated into 100 mL of Glc medium (pH 6.5) containing 60 mM glucose, 0.3% glutamate, 0.52% KH2PO4, 0.152% KCl, 0.05% adenine, and 0.15% methionine and cultured for 2 days at 90 rpm and 30 °C. For the induction, the strains were cultured in Sor medium (60 mM sorbitol and the same contents listed for Glc medium, except for glucose). Pellets were placed on glass slides and disentangled using a needle. The glass slides were observed using an HS All-in-one Fluorescence Microscope BZ-9000 (Keyenece, Osaka, Japan) with exposure for 1/350 s and a gain of 0 dB for light microscopy and exposure for 1/2 s and a gain of + 12 dB for fluorescence microscopy.
Western blotting
Conidia of sorA and sorB promoters::EGFP strains were cultured in 100 mL of Glc and Sor medium for 2 days at 90 rpm and 30 °C. Mycelium were harvested and dried. Mycelium (0.1 g) were dissolved in 300 μL of SDS-PAGE buffer and mixed and heated at 100 °C for 10 min. Samples were centrifuged for 5 min at 13,500 rpm, and supernatants were subjected to SDS-PAGE. Proteins were then transferred to PVDF membranes. EGFP was detected using anti-GFP rabbit polyclonal serum (Invitrogen), anti-IgG AP (Invitrogen), and the Proto BlotII AP system with stabilized substrate (Promega) according to the manufacturer’s instructions. For the analysis of other carbon source induction, each strain was cultured in the medium (contained 60 mM each carbon source and the same contents listed for Glc medium, except for glucose) for 2 days at 90 rpm and 30 °C. Mycelia were harvested and detected with the use of EGFP by the same procedure described above.
Results
Identification of sorbitol- and galactose-responsive genes
We performed a microarray analysis to identify A. oryzae genes that were highly expressed after switching from glucose media to a 1-h incubation with galactose and sorbitol induction. We obtained the expression profiles and compared the expression data following galactose and sorbitol incubation with the data of the mycelia grown in glucose (Fig. ). The gene expressions did not vary between the mycelia grown in galactose and those grown in glucose (Fig. (A)).
Fig. 1. Microarray analysis of sorbitol-responsive genes using GeneChip.
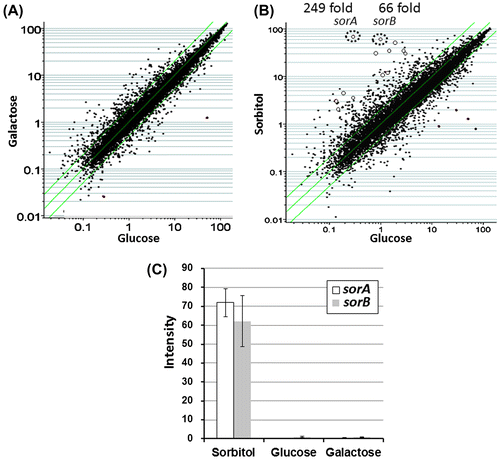
With galactose induction, no genes were expressed at a level more than 10-fold higher than that of glucose. However, when mycelia grown in sorbitol were compared with those kept in glucose, there were 13 genes that showed > 10-fold higher expression in sorbitol compared to that in glucose (Fig. (B)). We designated these induced genes sorA-M (Table ). Among the 13 genes with expression that was highly induced by sorbitol, sorA (AO090020000639) and sorB (AO090020000635) demonstrated more than 50-fold higher expression in sorbitol compared to that in glucose (Fig. (B) and Table ).
Table 3. Sorbitol-responsive genes in A. oryzae.
According to CAoGD (http://nribf21.nrib.go.jp/CFGD/), sorA and sorB are localized to the left arm of the sixth chromosome in A. oryzae strain RIB40. sorA encodes sugar transporter-like protein (stl1) and sorB encodes sorbitol dehydrogenase (xyl2). Confirming the microarray results, these two genes were not expressed in the mycelia grown in the media containing glucose or galactose (Fig. (C)). We selected the promoters for sorA and sorB for the promoter shutoff system development.
To determine the transcriptional start point for each gene, we performed 5′-RACE and found that the amplified length of each transcript was approximately 700 bp (sorA) and 1000 bp (sorB) (data not shown). The amplified fragments were cloned and sequenced with final transcriptional start points established between −57 and −8 (−57, −53, −32, −19, −17, −8) bp for sorA and at −36 bp for sorB (Supplementary Fig. S1).
To determine the physiological effects of each carbon source, we evaluated the colony growth in each media condition. The colony growth in glucose for 6 days (50.8 ± 0.53 mm) was greater than that in galactose (46.7 ± 0.87 mm) and sorbitol (45.7 ± 0.70 mm), but no morphological changes were noted. In addition, the expression of genes related to the glycolytic pathway, TCA cycle, electron transport chain, and pentose phosphate pathway did not change by more than twofold except for the expression of pfkA (phosphofructokinase) and fbpA (fructose bisphosphatase) in the glycolytic pathway when the carbon source was switched from glucose to sorbitol and galactose (Suppl. Fig. S2). Although the expression ratios of pfkA and fbpA changed, their signal intensities were still high (approximately 1000–4500), indicating that the above-mentioned pathways were still functioning. However, only the first reaction-related genes in the Leloir pathway (galactose metabolism) and the oxidative reduction pathway (sorbitol metabolism) were up-regulated under the sorbitol and galactose conditions, respectively. These results suggested that the carbon source did not significantly affect the physiological activities of A. oryzae.
Evaluation of sorbitol-responsive promoter induction in an engineered EGFP strain
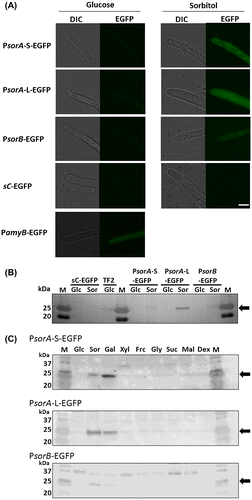
To evaluate the utility of the sorbitol-induced gene promoters, we constructed EGFP expression strains under the control of PsorA and PsorB. For PsorA, we constructed two strains with promoter lengths ranging from −1408 (PsorA-L) and −740 (PsorA-S) to −1. PsorA-L contained the full intergenic region (1408 bp) upstream of the sorA ORF. To design PsorA-S, we searched the CreA-,Citation20) Hap complex-,Citation21) and AreA-binding motif,Citation22) and TATA boxCitation23) in the promoter region and deduced the shortest region for repression (Suppl. Fig. S1). We also considered handling of the promoter for further use as an experimental tool. For PsorB, we constructed a strain with a promoter length ranging from −574 to −1. When the PsorA-S-EGFP, PsorA-L-EGFP, and PsorB-EGFP strains were cultured in glucose or sorbitol as the only carbon source, the EGFP fluorescence in the mycelia was observed in the sorbitol conditions, but not in the glucose conditions (Fig. (A)). The Western blot analysis was performed to confirm the EGFP expression in each condition. In each EGFP-inducible strain, EGFP was detected with the sorbitol conditions but not with the glucose conditions. The comparison of expression levels revealed that the PsorA-L-EGFP strain expressed the greatest amount of EGFP (Fig. (B)) and showed the highest fluorescence (Fig. (A)). The amyB promoter of A. oryzae has frequently been used as a conditional promoter, but it still shows some expression under repression conditions (Fig. (A) and (B)). The PsorA and PsorB tightly regulated the EGFP expression under the glucose-repressed conditions. To evaluate the induction ability of PsorA and PsorB using various carbon sources (galactose, xylose, fructose, glycerol, sucrose, maltose, and dextrin), we examined the EGFP expression by conducting a Western blot analysis (Fig. (C)). EGFP induction occurred in the sorbitol and galactose conditions in all three promoter strains but did not occur with any other carbon source. Thus, both PsorA and PsorB were inducible in sorbitol and in galactose but repressed in glucose, suggesting the use of these promoters as inducible promoters for studies of gene function.
Fig. 2. EGFP expression controlled by PsorA and PsorB, and the detection of EGFP in media containing various carbon sources by Western blotting.
An A. oryzae gene sorbitol promoter shutoff system
We next applied this sorbitol promoter system to regulate the A. oryzae original gene. brlA is a master regulator governing conidia formation; an A. oryzae brlA deletion strain shows the loss of conidia formation.Citation24) We constructed strains with brlA expression under the control of PsorA or PsorB (Suppl. Fig. S3B). Under the glucose-repressed conditions, both the PsorA-brlA and PsorB-brlA strains showed repressed conidia formation (Fig. ). Conversely, under the sorbitol induction conditions, both strains showed normal conidia formation. We then evaluated the conidia formation rate to confirm the number of conidia formed in each condition. In the glucose-containing media, both strains showed decreased conidia formation, and the control strains showed no change. These findings suggested that this promoter shutoff system can also be used for the evaluation of A. oryzae original gene regulation.
Fig. 3. Conidiation as controlled by PsorA-S and PsorB promoters. Both strains demonstrated normal conidiation under the sorbitol induction growth conditions, whereas reduced phialide formation and conidiation growth were observed under the glucose conditions. The right panel shows the Z-stack image of each plate. Scale bar: 200 μm. Values on the image indicate the conidia formation rate [conidia count ( × 105/cm2) ± SD].
![Fig. 3. Conidiation as controlled by PsorA-S and PsorB promoters. Both strains demonstrated normal conidiation under the sorbitol induction growth conditions, whereas reduced phialide formation and conidiation growth were observed under the glucose conditions. The right panel shows the Z-stack image of each plate. Scale bar: 200 μm. Values on the image indicate the conidia formation rate [conidia count ( × 105/cm2) ± SD].](/cms/asset/2766b085-78c7-4c7e-a8a0-899d5fa817b5/tbbb_a_1189313_f0003_oc.gif)
Use of the promoter shutoff system for essential gene analysis
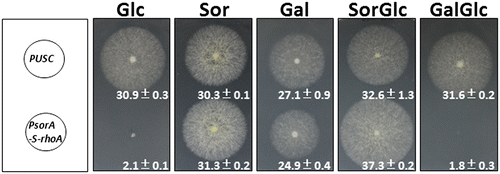
To apply this sorbitol-inducible promoter shutoff system to the functional analysis of an essential gene, we constructed an inducible PsorA-rhoA strain. RhoA is a regulator of beta-1,3-glucan synthase (FksA), an essential component in cell wall synthesis that is required for normal mycelia growth.Citation25) In A. niger, the deletion of rhoA is reportedly lethal and only rhoA heterokaryon strains survive.Citation26) We selected PsorA for an essential gene analysis with an expectation of tight regulation because the PsorA promoter region harbors more putative CreA-binding motifs,Citation20) and these motifs bind a carbon catabolite repressor. We constructed a PsorA-S-rhoA strain (Suppl. Fig. S3C) and evaluated its growth inhibition when mycelia were grown with different carbon sources. The PsorA-S-rhoA strain showed complete growth inhibition under the glucose-repression condition and normal growth under the sorbitol and galactose conditions (Fig. ). On the sorbitol and glucose-containing plates, the PsorA-S-rhoA strain showed normal growth, indicating that sorbitol works as an inducer. However, on the galactose and glucose-containing plates, the PsorA-S-rhoA strain showed complete growth inhibition, indicating that carbon catabolite repression occurred under this condition. These observations suggested the ability to tightly regulate rhoA gene expression, and these results indicated that this newly developed sorbitol-responsive promoter shutoff system will have utility in the essential gene analysis of filamentous fungi.
Fig. 4. Promoter shutoff analysis of the essential rhoA gene as controlled with PsorA-S.
Discussion
In this study, we aimed to identify sorbitol-inducible genes in A. oryzae and to develop a new promoter shutoff system for the evaluation of an essential gene. There are several conditional promoters in A. oryzae, including PalcA, PamyB, and PthiA, but their use has limitations. We developed the promoter system described herein to overcome these limitations and to create truly inducible promoters in A. oryzae. Because a carbon source exchange often has pleiotropic effects, we focused on the galactose and sorbitol metabolic pathways as they are expected to enter the glycolytic pathway through a few metabolic steps, thus leading to fewer physiological effects within filamentous fungi.
In the oxido-reductive D-galactose catabolic pathway, D-galactose is deoxidized to galactitol by XyrA and then dehydrogenized to L-xylo-3-hexulose by LadB, with eventual conversion to D-sorbitol. D-sorbitol is then dehydrogenized to D-fructose by SdhA, which enters into the glycolytic pathway in A. niger.Citation12) The results of our microarray analysis following galactose and sorbitol induction revealed no significant changes in gene expression except for changes in some of the genes such as sorA and sorB. In particular, when galactose and glucose were compared, no genes were induced at levels of 10-fold or higher by galactose. However, incubation with sorbitol led to the detection of 13 sorbitol-induced genes. The sorbitol-inducible sorA to sorM genes may be directly involved in sorbitol metabolism. Based on our search of the PRINTS database (http://www.bioinf.manchester.ac.uk/dbbbrowser/PRINTS/index.php), SorA harbors a sugar transporter signature (PR00171) motif based on a search of PRINTS and may function as a sorbitol transporter. In addition to the SorB ortholog in A. niger, SdhA functions as D-sorbitol dehydrogenase.Citation12) The A. niger strain ΔsdhA showed a growth defect in media containing sorbitol.Citation12) We observed that ΔsorA and ΔsorB strains of A. oryzae also showed growth defects in sorbitol (data not shown). We therefore expected both genes to be induced by sorbitol, and we subjected them to the further development of a unique new promoter system.
By comparing the A. oryzae growth in glucose as an optimal carbon source with the growth in sorbitol and galactose, we observed that a slight delay occurred when mycelia were grown in galactose (Fig. ). However, the cell morphology did not change under any growth condition. In fact, the microarray results demonstrated that each of the conditions did not cause much of a change in gene expression (Fig. (B)). The expression of genes related to the major energy metabolism did not change by more than 2-fold when the carbon source was switched from glucose to sorbitol and galactose (Suppl. Fig. S2). Only the first reaction-related genes in the Leloir pathway (galactose metabolism) and the oxidative reduction pathway (sorbitol metabolism) were up-regulated under the sorbitol and galactose conditions, respectively. Therefore, we surmised that the change in carbon source resulting in induction may not affect the physiology or whole gene expression within mycelium.
When PsorA and PsorB are used for gene induction, they may affect mycelial growth: This is because the expressions of sorA and sorB are reduced and the sorbitol metabolism is decreased by competing with the transcription factor of these genes. However, the PsorA regulation of the expressions of egfp (data not shown), brlA (Fig. ), and rhoA (Fig. ) expression did not demonstrate any special phenotypic differences such as changes in the growth or morphology of mycelia growth in sorbitol-containing media. Thus, this novel promoter shutoff system can be safely used for the analyses of gene function by induction.
To test this induction in other carbon sources (i.e. galactose, xylose, fructose, glycerol, sucrose, maltose, and dextrin), we conducted EGFP reporter assays with each carbon source and assessed the EGFP by Western blotting (Fig. (C)). We were unable to detect EGFP regulated by any of the three promoter constructs under most of the carbon source conditions; however, EGFP was clearly detected in the galactose- and sorbitol-containing media. The expressions of sorA and sorB were not detected by our DNA microarray analysis under the galactose induction conditions, although we detected EGFP induction in galactose. Moreover, in the PsorA-S-EGFP strain, we detected increased EGFP in galactose compared to that in sorbitol. We noticed that a similar induction also occurred in the PsorA-S-rhoA strain, which survived when grown in galactose or sorbitol (Fig. ). We speculate that the galactose induction of sorA and sorB gene expression was slower than the sorbitol induction. sdhA was expressed in A. niger after 2 h of induction in sorbitol and after 8 h in galactose.Citation12) The mechanism of galactose induction with PsorA and PsorB may not be direct; this requires further analysis.
For repression of PsorA and PsorB, the carbon catabolite repressor CreA may bind to the promoter and repress gene expression as well as the alcA promoter. We thus searched the deduced carbon catabolite element (5′-SYGGRG-3′) in the PsorA and PsorB sequence (Suppl. Fig. S1A, B). We found 10 deduced CREA-binding sites in PsorA –L, and four such sites in PsorB, and in the glucose condition, the expression of sorA gene was lower than that of sorB (Fig. (B) and (C)). We thus expect more a stronger repression of PsorA than PsorB, and we attempted to construct a PsorA-S-rhoA strain for evaluating promoter shutoff ability of PsorA-S. We observed that the repression of rhoA controlled with PsorA-S was different between the sorbitol-and-glucose mixed condition and the galactose-and-glucose mixed condition (Fig. ). Tight carbon catabolite repression occurred for the rhoA expression in the glucose-and-galactose mixed condition, but induction still occurred in the glucose-and-sorbitol mixed condition, suggesting that the induction mechanism of PsorA might be different for galactose and sorbitol. When applying this system for essential gene functional analyses, sorbitol can be used as an inducer for gene induction in the glucose-repression condition and glucose can be used as a repressor for promoter shutoff in the galactose induction condition.
This promoter shutoff system can be directly applied to fermentation engineering studies. Although its ability to induce expression is low (Fig. (B)), this system is repressed by glucose, which is an inexpensive carbon source. Moreover, this system has potential benefits in the evaluation of the expression of potentially toxic products of filamentous fungi because of its tight regulation of cell function. The sorbitol metabolism appears to be a fundamental pathway within filamentous fungi, and if sorbitol is used as the only carbon source, this sorbitol promoter shutoff system can become a commonly used experimental tool. The A. oryzae genome sequence revealed that approximately one-half of the genes that have been identified within the genome still have unknown functions.Citation27) We have begun using this promoter shutoff system to analyze essential genes with unknown functions. We anticipate that this promoter shutoff system will become a practical molecular biological tool that will contribute to the functional studies of filamentous fungi.
Author contributions
KO, MK, and KI designed the experiments; KO, ST, RH, RT, and KI performed the experiments; KO, MK, HF, and KI analyzed the data; and KO and KI wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was partially supported by the grant from the Japan Sake and Shochu Makers Association.
Supplementary material
The supplementary material for this paper is available online at http://dx.doi.org/10.1080/09168451.2016.1189313.
TBBB_1189313_Supplementary_Captions.docx
Download MS Word (44.1 KB)TBBB_1189313_S3.tif
Download TIFF Image (356.2 KB)TBBB_1189313_S2.tif
Download TIFF Image (126.6 KB)TBBB_1189313_S1.tif
Download TIFF Image (121.9 KB)Acknowledgment
The authors acknowledge Ryoko Hamada for her technical help with GeneChip microarray analysis.
Notes
Abbreviations: PsorA, sorA promoter; PsorB, sorB promoter; DIC, differential interference contrast; CAoGD, comprehensive Aspergillus oryzae genome database; EGFP, enhanced green fluorescence protein.
References
- Murakami H. The study of Koji. Brew. Soc. Japan. 1976;1:1–31 ( in Japanese).
- Zarrin M, Leeder AC, Turner G. A rapid method for promoter exchange in using recombinant PCR. Fungal Genet. Biol. 2005;42:1–8.10.1016/j.fgb.2004.10.002
- Romero B, Turner G, Olivas I, et al. Ramón De Lucas J. The Aspergillus nidulans alcA promoter drives tightly regulated conditional gene expression in Aspergillus fumigatus permitting validation of essential genes in this human pathogen. Fungal Genet. Biol. 2003;40:103–114.10.1016/S1087-1845(03)00090-2
- Tada S, Gomi K, Kitamoto K, et al. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli beta-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol. Gen. Genet. 1991;229:301–306.10.1007/BF00272170
- Shoji JY, Maruyama JI, Arioka M, et al. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 2005;244:41–46.10.1016/j.femsle.2005.01.014
- Ishida H, Matsumura K, Hata Y, et al. Establishment of a hyper-protein production system in submerged Aspergillus oryzae culture under tyrosinase-encoding gene (melO) promoter control. Appl. Microbiol. Biotechnol. 2001;57:131–137.
- Tsuboi H, Koda A, Toda T, et al. Improvement of the Aspergillus oryzae enolase promoter (P-enoA) by the introduction of cis-element repeats. Biosci. Biotechnol. Biochem. 2005;69:206–208.10.1271/bbb.69.206
- Ishida H, Hata Y, Kawato A, et al. Improvement of the glaB promoter expressed in solid-state fermentation (SSF) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2006;70:1181–1187.10.1271/bbb.70.1181
- Juvvadi PR, Kuroki Y, Arioka M, et al. Functional analysis of the calcineurin-encoding gene cnaA from Aspergillus oryzae: evidence for its putative role in stress adaptation. Arch. Microbiol. 2003;179:416–422.
- Som T, Kolaparthi VS. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol. Cell. Biol. 1994;14:5333–5348.10.1128/MCB.14.8.5333
- Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470.
- Koivistoinen OM, Richard P, Penttilä M, et al. Sorbitol dehydrogenase of Aspergillus niger, SdhA, is part of the oxido-reductive d-galactose pathway and essential for d-sorbitol catabolism. FEBS Lett. 2012;586:378–383.10.1016/j.febslet.2012.01.004
- Sambrook J, Russel D. Molecular cloning: a laboratory manual. 3rd ed. New York (NY): Cold spring harbor laboratory press; 2001.
- Kawauchi M, Nishiura M, Iwashita K. Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA. Eukaryot. Cell. 2013;12:1087–1096.10.1128/EC.00003-13
- Yamada O, Nan SN, Akao T, et al. dffA gene from Aspergillus oryzae encodes L-ornithine N-5-oxygenase and is indispensable for deferriferrichrysin biosynthesis. J. Biosci. Bioeng. 2003;95:82–88.10.1016/S1389-1723(03)80153-6
- Yamada O, Lee BR, Gomi K. Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci. Biotechnol. Biochem. 1997;61:1367–1369.10.1271/bbb.61.1367
- Maruyama JI, Kitamoto K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol. Lett. 2008;30:1811–1817.10.1007/s10529-008-9763-9
- Gomi, K; Iimura, Y; Hara S. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 1987;51:2549–2555.10.1271/bbb1961.51.2549
- Szewczyk E, Nayak T, Oakley CE, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006;1:3111–3121.
- Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415.
- Nagata O, Takashima T, Tanaka M, et al. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible Taka-amylase A gene. Mol. Gen. Genet. 1993;237:251–260.
- Ravagnani A, Gorfinkiel L, Langdon T, et al. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 1997;16:3974–3986.10.1093/emboj/16.13.3974
- Kuchrski R, Bartnik E. The TBP gene from Aspergillus nidulans-structure and expression in Saccharomyces cerevisiae. Molecularbiology. 1997;143:1263–1270.
- Yamada O, Lee BR, Gomi K, et al. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J. Biosci. Bioeng. 1999;87:424–429.10.1016/S1389-1723(99)80089-9
- Guest GM, Lin X, Momany M. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 2004;41:13–22.10.1016/j.fgb.2003.08.006
- Kwon MJ, Arentshorst M, Roos ED, et al. Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol. Microbiol. 2011;79:1151–1167.10.1111/mmi.2011.79.issue-5
- Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161.10.1038/nature04300
- Oda K, Kakizono D, Yamada O, et al. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006;72:3448–3457.10.1128/AEM.72.5.3448-3457.2006
