Abstract
Under liquid culture conditions, the hyphae of filamentous fungi aggregate to form pellets, which reduces cell density and fermentation productivity. Previously, we found that loss of α-1,3-glucan in the cell wall of the fungus Aspergillus nidulans increased hyphal dispersion. Therefore, here we constructed a mutant of the industrial fungus A. oryzae in which the three genes encoding α-1,3-glucan synthase were disrupted (tripleΔ). Although the hyphae of the tripleΔ mutant were not fully dispersed, the mutant strain did form smaller pellets than the wild-type strain. We next examined enzyme productivity under liquid culture conditions by transforming the cutinase-encoding gene cutL1 into A. oryzae wild-type and the tripleΔ mutant (i.e. wild-type-cutL1, tripleΔ-cutL1). A. oryzae tripleΔ-cutL1 formed smaller hyphal pellets and showed both greater biomass and increased CutL1 productivity compared with wild-type-cutL1, which might be attributable to a decrease in the number of tripleΔ-cutL1 cells under anaerobic conditions.
Graphical abstract
Summary of the growth characteristic and enzyme productivity in A. oryzae ags tripleΔ strain.
Many filamentous fungi, such as those in genus Aspergillus, secrete commercially valuable enzymes and metabolites and are therefore of high value to the fermentation industry.Citation1–3) Although filamentous fungi secrete higher levels of these valuable enzymes and metabolites than yeasts and bacteria, under liquid culture conditions the hyphae of filamentous fungi often clump together to form large pellets,Citation4) which prevents the establishment of high cell densities under liquid culture conditions. Therefore, preventing pellet formation is crucial for improving the productivity of commercial fermentation processes.
The aggregation of hyphae and formation of pellets is thought to be related to the biochemical properties of the outer layer of the hyphae,Citation5) which is composed mainly of cell wall polysaccharides and cell wall-associated proteins.Citation6) Therefore, further elucidation of the relationship between hyphal aggregation and the chemical components, particularly the cell wall polysaccharides, of the hyphal cell surface is needed.
Several chemical analyses of the components of the cell wall of the human pathogen Aspergillus fumigatus have been conducted.Citation7,8) The major components of the A. fumigatus cell wall are polysaccharides, which can be fractionated into those that are alkali soluble (AS) and those that are alkali insoluble (AI). The AS fraction mainly contains α-1,3-glucan with some galactomannan, whereas the AI fraction mainly contains galactomannan, chitin, and highly branched β-1,3-glucan with β-1,6 linkages (β-1,3/1,6-glucan).Citation7) In the hyphal cells of A. fumigatus grown under liquid culture conditions, the major component of the AI fraction is β-1,3/1,6-glucan, which forms a three-dimensional network with numerous non-reducing endsCitation7); chitin, galactomannan, and β-1,3/1,4-glucan are covalently bonded to the non-reducing ends of the β-1,3/1,6-glucan, producing a large hetero-polysaccharide network.Citation7) Consistent with these findings, Yoshimi et al. reported that the components of the cell wall of the model filamentous fungus Aspergillus nidulans are similar to those of A. fumigatus,Citation4) and Mizutani et al. reported that the components of the cell wall of the industrial filamentous fungus A. oryzae are similar to those of A. fumigatus.Citation9)
Cell wall α-1,3-glucan was first recognized as a virulence factor in pathogenic fungi such as the human pathogenic dimorphic yeast Histoplasma capsulatum,Citation10–12) the human pathogen A. fumigatus,Citation13,14) and the plant pathogenic filamentous fungus Magnaporthe grisea.Citation15,16) α-1,3-Glucan in the cell wall of pathogenic yeasts and filamentous fungi is thought to be distributed in the outer layer of the cell wall and to cover β-1,3-glucan or chitin to prevent immune recognition by host cells.Citation12,15)
We previously reported that transcription of the two genes encoding α-1,3-glucan synthase (i.e. agsA and agsB) in A. nidulans is mainly regulated via the cell wall integrity signaling pathway.Citation17) To determine the function of agsA and agsB other than as stealth factors against recognition by host cells, we constructed three strains of A. nidulans in which agsA, agsB, or both were disrupted (i.e. agsAΔ, agsBΔ, and agsAΔagsBΔ).Citation4) We found that the agsBΔ and agsAΔagsBΔ strains had no α-1,3-glucan in their cell wall, whereas the agsAΔ strain had the same amount of α-1,3-glucan in its cell wall as did the wild-type strain, suggesting that AgsB is the major α-1,3-glucan synthase in A. nidulans. Furthermore, whereas the hyphae of the wild-type and agsAΔ strains aggregated to form pellets under liquid culture conditions, the hyphae of the agsBΔ and agsAΔagsBΔ strains were dispersed.Citation4) Together, these results suggest that under liquid culture conditions α-1,3-glucan is a hyphal cell aggregation factor in A. nidulans.
In industrial fermentation using single-cell micro-organisms such as bacteria and yeasts, cultures of high cell density are necessary for high productivity. However, most industrial filamentous fungi, including aspergilli, form hyphal pellets under liquid culture conditions, leading to low cell density and limited productivity. Because hyphal cells of A. nidulans mutants lacking α-1,3-glucan in their cell wall are dispersed,Citation4) we hypothesized that this may also be true of other industrial fungi, such as A. oryzae, and therefore disruption of the genes involved in α-1,3-glucan biosynthesis represents a possible strategy for reducing the pellet formation and increasing cell density and fermentation productivity under liquid culture conditions.
In the present study, we constructed an A. oryzae mutant in which we disrupted all three genes encoding α-1,3-glucan synthases (i.e. agsA, agsB, agsC; A. oryzae tripleΔ) and examined the effects of this disruption on pellet formation. Recently, two novel α-1,3-glucan synthesis-related genes, amyD and amyG, which putatively encode a GPI-anchored glycosidase and an intracellular glycosidase, respectively, were reported in A. nidulans.Citation18) These two glycosidase-encoding genes together with agsB form a gene cluster, the transcription of which is coordinately regulated in A. nidulans.Citation18) Therefore, we also constructed an A. oryzae agsAΔagsBΔagsCΔamyDΔamyGΔ quintuple disruptant (A. oryzae quintupleΔ). To examine enzyme productivity, we transformed a single copy of the A. oryzae cutL1 gene encoding the cutinase CutL1, a model secretory enzyme that degrades biodegradable aliphatic polyester,Citation19) into the wild-type, tripleΔ, and quintupleΔ strains.
Materials and methods
Strains and growth media
Strains used in this study are listed in Table . A. nidulans ABPU1 (biA1, pyrG89, wA3, argB2, pyroA4) with ligD∆ (ligD∆::ptrA)Citation4) and A. oryzae NS4 (sC−, niaD−) with ligDΔ (ligDΔ::sC, adeAΔ::ptrA) were used for all genetic manipulations.Citation20) A. nidulans agsAΔagsBΔ, a strain in which the two genes encoding α-1,3-glucan synthases (i.e. agsA, agsB) were disrupted, was produced as previously described.Citation4) All A. nidulans strains were cultivated in Czapek-Dox (CD) medium containing 0.6% (70 mM) NaNO3, 0.052% KCl, 0.152% KH2PO4, 0.059% MgSO4·7H2O, 0.1% trace elements solution [0.1% FeSO4·7H2O, 0.88% ZnSO4·7H2O, 0.04% CuSO4·5H2O, 0.015% MnSO4·4H2O, 0.01% Na2B4O7·10H2O, 0.005% (NH4)6Mo7O24·4H2O], and 2% glucose. The CD medium was adjusted to pH 6.5. To fulfill the auxotrophic requirements of A. nidulans, 200 μg of arginine/mL, 0.02 μg of biotin/mL, 0.5 μg of pyridoxine/mL, 1.22 mg of uridine/mL, and 1.12 mg of uracil were added to the CD medium.
Table 1. Strains used in this study.
A. oryzae niaD− strains were cultivated in CDE medium, which was CD medium in which the nitrogen source was 70 mM of sodium hydrogen L(+)-glutamate monohydrate instead of 70 mM NaNO3. A. oryzae niaD (CutL1-expressing) strains were cultured in CD medium described above. The strain of A. oryzae requiring adenine was cultivated in CDE medium supplemented with 0.01% adenine (CDEA). To construct A. oryzae tripleΔ (agsAΔagsBΔagsCΔ) strain, we used the adeA-marker recycle method as described below. The adeA marker of the gene disruption cassette integrated in the chromosome of A. oryzae was excised by Cre-loxP system,Citation21,22) resulting in the generation of adeA− strains that were grown in CDEA medium.
Conidia of A. oryzae used to inoculate flask cultures were isolated from cultures grown on malt medium containing 9% malt extract (Becton Dickinson and Company, Sparks, USA), 0.5% yeast extract (Becton Dickinson and Company), and 0.1% trace elements solution. YPD medium containing 2% peptone (Becton Dickinson and Company), 1% yeast extract, and 2% glucose was used as the medium for the flask cultures used to analyze the growth characteristics of the strains. YPM medium containing 2% peptone, 1% yeast extract, and 2% maltose was used as the medium for the flask cultures to analyze the secretion of CutL1. YPDS medium containing 6% peptone, 1% yeast extract, 6% glucose, and 20 mM succinate buffer (pH 7.0) was used for the flask cultures to analyze CutL1 secretion under conditions of suppressed amylase production.
Construction of A. oryzae tripleΔ and quintupleΔ strains
The disruption cassette for each of the three ags genes was constructed as shown in Fig. S1(A). The pYES2-based vectorCitation17) containing the disruption cassette for each ags gene was constructed with following amplicons (A, L, R, B). Amplicon A: the yeast-Escherichia coli shuttle vector pYES2 was digested with BamHI and EcoRI. Amplicon L: the DNA fragment corresponding to −1500 to −500 bp upstream the translation initiation site was amplified with wild-type genomic DNA template and primers (agsA-LU [or agsB-LU or agsC-LU] and agsA-LL-loxP [or agsB-LL-loxP or agsC-LL-loxP]) by polymerase chain reaction (PCR). Amplicon R: the DNA fragment corresponding to +2500–3500 bp downstream the translation initiation site was amplified with wild-type genomic DNA and the primers (agsA-RU-loxP [or agsB-RU-loxP or agsC-RU-loxP] and agsA-RL [or agsB-RL or agsC-RL]) by PCR. Amplicon B: A. nidulans adeA marker and Cre-expression cassette encompassed with two loxP fragment were amplified with pAAAXP-Cre (Zhang et al., in submission) as a template and the primers (adeA-Cre-loxP-Fw and adeA-Cre-loxP-Rv) by PCR. The amplicons A, L, R, and B were fused in order to obtain each ags gene disruption vector (Fig. S1(A)) by homologous recombination in S. cereviae BY4741.Citation23) The disruption cassette fragment was amplified with the disruption vector for each ags gene by PCR. Primers for PCR are listed in Table S1.
The agsA gene in A. oryzae NS4 (sC−, niaD−) with ligDΔ (ligDΔ::sC, adeAΔ::ptrA) was disrupted with the agsA-disruption cassette and the agsA disrutants were selected as adeA+ transformants and confirmed by Southern blot analysis. The adeA marker gene in the agsA disruptant was excised by induction of Cre-recombinase with 1% xylose (instead of glucose) in the CDEA medium (Fig. S1(A)). The agsAΔagsBΔ strains were obtained from the agsAΔ strain by using the agsB disruption cassette containing adeA marker. The agsAΔagsBΔagsCΔ strains were constructed from the double mutant by using the adeA marker recycle method.
To construct the dual disruption cassette for the amyD and amyG genes, gene fragments containing the 5′ non-coding region of amyD (amplicon 1), the 3′ non-coding region of amyG (amplicon 2) derived from the A. oryzae genomic DNA template, and the adeA gene (amplicon 3) from the plasmid TOPO-2.1-adeA (described below) were amplified by means of PCR (Table S1). Amplicon 1 was amplified with the primers amyD-LU and amyD-LL + Ade, amplicon 2 with the primers amyG-RU + Ade and amyG-RL, and amplicon 3 with the primers amyD-AU and amyG-AL. The primers amyD-LL + Ade, amyG-RU + Ade, amyD-AU, and amyG-AL were chimeric oligonucleotides; each contained a reverse-complement sequence for PCR fusion. The PCR products were gel-purified and then used as the substrates for a second round of PCR using the primers amyD-LU and amyG-RL to fuse the three fragments. The resulting major PCR product was gel-purified and used to transform A. oryzae tripleΔ (Fig. S1(B)). Disruption of the amyD and amyG genes was confirmed by means of Southern blot analysis (Fig. S1(C)).
The plasmid TOPO-2.1-adeA was constructed by following methods. The DNA fragment containing A. oryzae adeA marker gene was amplified with A. oryzae wild-type genomic DNA as a template by PCR. The amplified fragment containing the adeA gene was cloned into pCR2.1-TOPO vector (Invitrogen, Tokyo, Japan) by TA cloning according to the manufacturer’s instructions. This plasmid was used for the PCR template to construct the amyD and amyG gene disruption cassette and for complementation of A. oryzae adeA− strains.
Scanning electron microscopy
Samples for scanning electron microscopy analysis were prepared by following the method of Mizutani et al. with slight modification.Citation24) Colonies of A. oryzae wild-type, tripleΔ, and quintupleΔ strains grown on CDE agar plates were fixed, dehydrated, and lyophilized in tert-butanol. The lyophilized colonies were coated with platinum–vanadium and observed under a Hitachi SU8000 scanning electron microscope (Hitachi, Tokyo, Japan) at an accelerating voltage of 3 kV.
Construction of CutL1-expressing strains of A. oryzae
To allow comparative analysis of enzyme productivity, we constructed A. oryzae wild-type, tripleΔ, and quintupleΔ strains overexpressing cutinase CutL1 as a model secretory enzyme (i.e. A. oryzae wild-type-cutL1, tripleΔ-cutL1, and quintupleΔ-cutL1). To construct the CutL1 overexpression system, plasmid pNGA-gla-cutCitation25) expressing the A. oryzae cutL1 gene under the control of the A. oryzae glaA142 promoter was transformed into A. oryzae wild-type, tripleΔ, and quintupleΔ by following the methods of Gomi et al.Citation26) CutL1-overexpressing transformants were selected by using the niaD gene as a selectable marker. We confirmed that a single copy of the overexpression construct was integrated at the niaD locus by means of Southern blot analysis (Fig. S2).
Fractionation of cell wall components and quantitative analysis of the carbohydrate composition of the cell wall
Conidia (final concentration, 1 × 105/mL) of individual strains were inoculated into 200 mL of YPD liquid medium and rotated at 160 rpm at 30 °C for 48 h in a 500-mL Erlenmeyer flask. Cultured mycelia were collected by filtration through Miracloth (Merck Millipore, Darmstadt, Germany), washed twice with 20 mL of distilled water, and lyophilized. Lyophilized mycelia were pulverized by using a MM400 bench-top mixer mill (Retsch, Haan, Germany), and 0.5 g of the resulting powder was suspended in 40 mL of 0.1 M Na phosphate buffer (pH 7.0). Fractionation of the cell wall components by alkali-treatment was performed as described previously.Citation4) Carbohydrate composition of each cell wall fraction was quantitatively determined by following the methods of Yoshimi et al.Citation4)
Analysis of the growth characteristics of A. nidulans and A. oryzae under liquid culture conditions
Conidia (final concentrations, 1 × 105/mL) of A. nidulans wild-type or agsAΔagsBΔ were inoculated into 200 mL of CD liquid medium in 500-mL Erlenmeyer flasks and rotated at 160 rpm at 30 °C for 60 h. Mycelia were then collected and dried as described below. Conidia (final concentrations, 1 × 105/mL) of A. oryzae wild-type, tripleΔ, quintupleΔ, wild-type-cutL1, tripleΔ-cutL1, or quintupleΔ-cutL1 were inoculated into 200 mL of YPDS liquid medium and rotated at 160 rpm at 30 °C for 72 h in 500-mL Erlenmeyer flasks. Samples (5 mL) were withdrawn from the culture broth every 12 h and filtered through Miracloth to isolate the mycelia. The mycelia were washed twice with 20 mL of distilled water, dried at 70 °C for 24 h, and then weighed. Filtrates were stored in glass vials in a freezer (−20 °C) until use. Immediately after defrosting, the pH was measured and the concentrations of glucose and CutL1 (see below) in the filtrate were determined. The amount of glucose remaining in the culture broth was measured by using a glucose C II test kit (Wako, Osaka, Japan). The mean diameter of the hyphal pellets was determined by measuring 10 randomly selected pellets under a stereomicroscope (M125; Leica Microsystems, Wetzlar, Germany).
Assay of enzyme productivity under liquid culture conditions
Conidia (final concentration, 1 × 105/mL) of A. oryzae wild-type-cutL1, tripleΔ-cutL1, and quintupleΔ-cutL1 were inoculated into 50 mL of YPM liquid medium and rotated at 100 rpm at 30 °C for 24 h in 200-mL Erlenmeyer flasks. The culture broth was then filtered through Miracloth to remove the mycelia, and the filtrates were collected and stored as described above. To quantify the amounts of the proteins secreted, 100% (w/v) trichloroacetic acid was added to 200 μL of filtrate to precipitate the proteins. The precipitated proteins were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 17.5% SDS polyacrylamide gel) and stained with Coomassie Brilliant Blue. Gel images were captured by using a flatbed scanner (GT X-750; Seiko Epson Corp., Nagano, Japan) and the amount of CutL1 secreted in the broth was quantified densitometrically by using the United States National Institutes of Health’s ImageJ software (http://rsb.info.nih.gov/nih-image/about.html).
To further examine the details of CutL1 productivity, A. oryzae wild-type-cutL1 and tripleΔ-cutL1 strains were cultured for 72 h in YPDS medium containing 6% glucose to suppress amylase production. Conidia (final concentration, 1 × 105/mL) of each strain were inoculated into 200 mL of YPDS medium in 500-mL Erlenmeyer flasks and rotated at 120 rpm at 30 °C for 72 h. An aliquot (5 mL) of the culture broth was withdrawn, and the concentration of secreted CutL1, pH, amount of glucose remaining in the filtrate, and mycelial dry weight, were determined. The amount of glucose remaining in the filtrate was determined by using the phenol/H2SO4 method.Citation27,28)
Results
Growth characteristics of A. nidulans wild-type and agsAΔagsBΔ and of A. oryzae tripleΔ and quintupleΔ
Yoshimi et al. previously reported that the hyphae of A. nidulans agsBΔ and agsAΔagsBΔ, which both lack α-1,3-glucan in their cell wall, were well dispersed under liquid culture conditions.Citation4) Here, we compared the dry weight of mycelia of A. nidulans wild-type and agsAΔagsBΔ grown in CD liquid medium and found that the dry weight of A. nidulans agsAΔagsBΔ mycelia was 1.5-fold higher than that of the wild-type mycelia (Fig. S3), which suggests that the increased hyphal dispersion in A. nidulans agsAΔagsBΔ resulted in greater biomass production under liquid culture conditions. To investigate whether removal of α-1,3-glucan from the cell wall of the industrial fungus A. oryzae would also result in increased hyphal dispersion and an increase in biomass, we constructed a strain of A. oryzae in which the three genes encoding α-1,3-glucan synthases (i.e. agsA, agsB, agsC) were genetically disrupted (Fig. S1(A)).
To observe the mycelial growth of the A. oryzae strains on plate medium, conidia (1 × 104/mL) of the wild-type and tripleΔ strains were inoculated at the center of CDE agar plates and cultured at 30 °C for four days. Both strains showed normal radial growth and conidiation (data not shown). To compare the morphology of the wild-type, triple∆, and quintuple∆ strains, we observed the conidiophores and mycelia of these strains under a scanning electron microscope. No significant differences in the morphologies of the conidiophores and mycelia were observed among the three strains (Fig. S4). These results suggest that deletion of the three ags genes did not affect the growth of A. oryzae under plate culture conditions. This result is consistent with that previously reported for A. nidulans agsBΔ and agsAΔagsBΔ, which were also found to grow normally under plate culture conditions.Citation4)
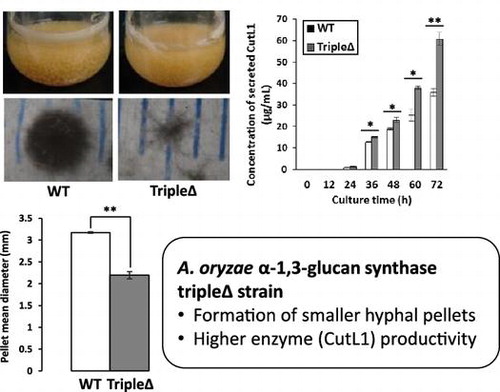
To examine the characteristics of mycelial growth in A. oryzae wild-type and tripleΔ under liquid culture conditions, conidia were inoculated into YPD liquid medium and rotated (160 rpm) at 30 °C for 48 h. At 36 h, the mean diameter of the hyphal pellets of A. oryzae tripleΔ (mean diameter, 2.2 mm) was 30% smaller than that of the hyphal pellets of wild-type A. oryzae (mean diameter, 3.2 mm) (p < 0.01; Fig. (A) and (B)). This suggests that loss of α-1,3-glucan in the cell wall caused by disruption of the three ags genes led to a weakening of the adhesive forces among the hyphae of A. oryzae tripleΔ. No differences in the branching of hyphae or the morphology of the hyphal tip were detected between the A. oryzae wild-type and tripleΔ strains in the microscopic assessment (data not shown). Similarly, A. nidulans wild-type and agsA∆agsB∆ also have comparable microscopic hyphal morphologies.Citation4)
Fig. 1. Phenotypes of Aspergillus oryzae agsAΔagsBΔagsCΔ triple mutant (tripleΔ) and agsAΔagsBΔagsCΔamyDΔamyGΔ quintuple mutant (quintupleΔ) under liquid culture conditions.
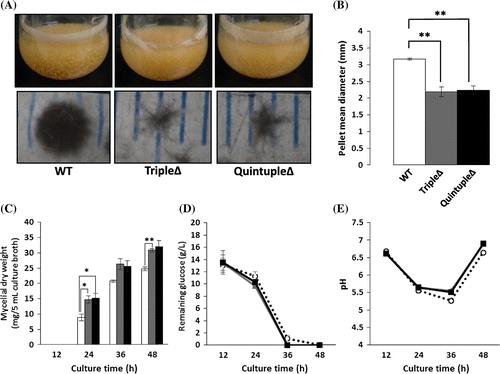
Whereas the hyphae of A. nidulans agsAΔagsBΔ were well dispersed under liquid culture conditions, those of A. oryzae tripleΔ were not and small pellets still formed. We hypothesized that the cells must still be producing a small amount of α-1,3-glucan and that it was causing the formation of the small hyphal pellets in the tripleΔ strain. He et al.Citation18) recently reported that A. nidulans amyD and amyG, which encode a GPI-anchored glycosidase and an intracellular glycosidase, respectively, were involved in the biogenesis of α-1,3-glucan in A. nidulans. Therefore, to try to further abolish the production of α-1,3-glucan in A. oryzae we constructed a strain in which the amyD and amyG genes were also disrupted (i.e. agsAΔagsBΔagsCΔamyDΔamyGΔ); however, the quintupleΔ strain still formed small hyphal pellets under liquid culture conditions (Fig. (A)).
Further characterization showed that the mycelial dry weights of A. oryzae tripleΔ and quintupleΔ were both greater than that of A. oryzae wild-type at 12–48 h of culture (Fig. (C)) and also that the two mutant strains consumed glucose slightly faster than did the wild-type strain (Fig. (D)). The pH of the culture broth in which tripleΔ and quintupleΔ was cultured was slightly higher than that of the culture broth in which the wild-type strain was cultured at 24–48 h of culture (Fig. (E)). Since the smaller pellets of A. oryzae tripleΔ and quintupleΔ (Fig. (B)) produced a larger total hyphal pellet surface area than that of the wild-type strain, fewer hyphal cells in the mutant strains would have been subject to anaerobic conditions. Thus, the larger surface area of the hyphal pellets of A. oryzae tripleΔ and quintupleΔ mean that these strains may be useful for improving the productivity of fermentation processes.
Analysis of the monosaccharide composition of the A. oryzae cell wall
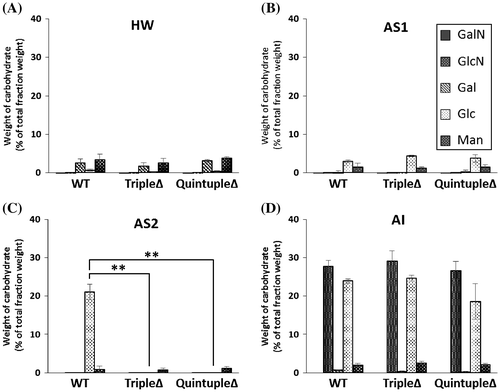
To examine the composition of the cell wall in A. oryzae wild-type, tripleΔ, and quintupleΔ, lyophilized hyphal cells of each strain were fractionated according to their solubility in alkali, and each fraction was hydrolyzed to determine the monosaccharide composition in the fraction. It has been reported that in A. fumigatus and A. nidulans the AS fraction of the cell wall contains mainly α-1,3-glucan with some galactomannan,Citation4,7) and that the AI fraction is composed of chitin, β-1,6-branched β-1,3-glucan, and galactomannan.Citation4,8) In the present study, the components of the cell wall from each strain were separated into four fractions: hot-water-soluble fraction (HW), water-soluble components after dialysis of the AS fraction (AS1), AS fraction (AS2), and AI fraction (AI), and the weight ratios of the fractions were then calculated (Table S2). The total recovery rate of the four fractions (HW + AS1 + AS2 + AI) was 55–65% of the total weight of lyophilized mycelia derived from each strain.
In A. oryzae wild-type, tripleΔ, and quintupleΔ, the HW fraction mainly contained galactose and mannose (Fig. (A)) and the AS1 fraction mainly contained glucose and mannose (Fig. (B)). The AS2 fraction derived from A. oryzae wild-type mainly contained glucose (approx. 20% of the AS2 fraction) with a small amount of mannose; however, those derived from A. oryzae tripleΔ and quintupleΔ contained significantly less glucose (p < 0.01; Fig. (C)). The AI fraction derived from each strain contained glucose, glucosamine, and mannose. The glucose and glucosamine contents of the AI fraction derived from each strain were around 28 and 20% of the total dry weight of the AI fraction, respectively (Fig. (D)). Together, these results show that although the ags genes play a major role in α-1,3-glucan biosynthesis in A. oryzae, the amyD and amyG genes do not. The overall composition of the cell wall in the amyDΔamyGΔ strain was very similar to that of the wild-type strain, and the amyDΔamyGΔ disruptant formed hyphal pellets of similar size to those of the wild-type strain (data not shown).
Fig. 2. Compositions of the monosaccharide fractions of the cell wall of Aspergillus oryzae wild-type (WT), agsAΔagsBΔagsCΔ triple mutant (tripleΔ), and agsAΔagsBΔagsCΔamyDΔamyGΔ quintuple mutant (quintupleΔ).
Cutinase productivity in A. oryzae tripleΔ-cutL1 and quintupleΔ-cutL1 under liquid culture conditions
To examine the growth characteristics of A. oryzae tripleΔ and quintupleΔ in which the cutL1 gene was overexpressed by the glaA142 promoterCitation29) (i.e. tripleΔ-cutL1 and quintupleΔ-cutL1 strains), A. oryzae wild-type-cutL1, tripleΔ-cutL1, and quintupleΔ-cutL1 were cultured for 24 h in YPM liquid medium containing maltose as an inducer of the A. oryzae glaA142 promoter. A. oryzae tripleΔ-cutL1 and quintupleΔ-cutL1 formed significantly smaller hyphal pellets than did the wild-type-cutL1 strain in YPM liquid medium (p < 0.01; Fig. (A) and (B)), which is consistent with the sizes of the pellets formed by A. oryzae wild-type, tripleΔ, and quintupleΔ in YPM liquid medium (data not shown). The mycelial dry weights of the tripleΔ-cutL1 and quintupleΔ-cutL1 strains were approximately twice that of wild-type-cutL1 (p < 0.01; Fig. (C)).
Fig. 3. CutL1 productivity of Aspergillus oryzae wild-type (WT), agsAΔagsBΔagsCΔ triple mutant (tripleΔ), and agsAΔagsBΔagsCΔamyDΔamyGΔ quintuple mutant (quintupleΔ) under liquid culture conditions.
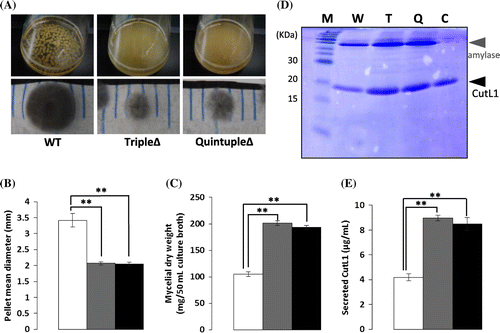
The enzyme productivity of A. oryzae wild-type-cutL1, tripleΔ-cutL1, and quintupleΔ-cutL1 was measured by means of SDS-PAGE analysis of CutL1 (21.6 kDa) secreted into culture broth (Fig. (D) and (E)). After 24 h of culture in YPM, the amount of CutL1 secreted by the tripleΔ-cutL1 and quintupleΔ-cutL1 strains was approximately twice that produced by the wild-type-cutL1 strain (p < 0.01; Fig. (E)).
To confirm that the improvement in protein productivity was attributable to the lack of α-1,3-glucan in the cell wall and the resulting small pellet size, we assessed the relationship between protein productivity and pellet size, controlled for inoculum size, under liquid culture conditions (Fig. S5). Conidia (final concentration, 1 × 103, 105, or 107/mL) of A. oryzae wild-type-cutL1 and tripleΔ-cutL1 strains were inoculated into 50 mL of YPM medium and rotated at 100 rpm at 30 °C for 24 h. In the cultures inoculated with 1 × 107/mL conidia, the wild-type-cutL1 strain produced pellets with a mean diameter of 1.9 mm, whereas in the culture inoculated with 1 × 105/mL, the wild-type-cutL1 strain produced pellets with a mean diameter of 3.4 mm (p < 0.05). The mean diameter of the pellets produced by the tripleΔ-cutL1 strain in the culture inoculated with 1 × 105/mL conidia (2.5 mm) was smaller than that of the pellets produced by the wild-type-cutL1 strain in the culture inoculated with 1 × 105/mL conidia (3.4 mm) (p < 0.05). The tripleΔ-cutL1 strain formed few hyphal pellets in the culture inoculated with 1 × 107/mL conidia. The amount of CutL1 secreted by the wild-type-cutL1 strain in the culture inoculated with 1 × 107/mL conidia was similar to that secreted by the tripleΔ-cutL1 strain in the culture inoculated with 1 × 105/mL conidia, but larger than the amount secreted by the wild-type strain in the culture inoculated with 1 × 105/mL conidia (Fig. S5).
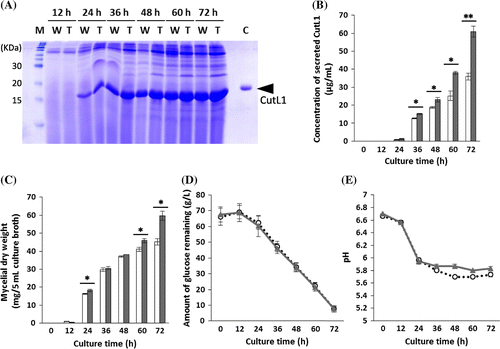
Because the phenotype of A. oryzae tripleΔ-cutL1, including CutL1 productivity, was very similar to that of the quintupleΔ-cutL1 strain, we further compared the CutL1 productivity of the tripleΔ-cutL1 strain with that of the wild-type-cutL1 strain by using liquid medium containing 6% glucose to mimic industrial culture conditions in which culture media often contain high concentrations of glucose or fructose. Samples of culture broth were withdrawn from the flasks every 12 h and the concentration of CutL1, mycelial dry weight, pH, and the amount of glucose remaining in the culture broth were determined. The amount of CutL1 secreted by the tripleΔ-cutL1 strain was approximately 150% of that secreted by the wild-type-cutL1 at 60 h (p < 0.05) and 170% of that secreted by the wild-type-cutL1 at 72 h (p < 0.01) (Fig. (A) and (B)). Although the growth rate (mean 8.2 mg/24 h) of the wild-type-cutL1 strain was decreased at 48 h, that (mean 21.7 mg/24 h) of tripleΔ-cutL1 was not changed even at 72 h (p < 0.01) (Fig. (C)). These results suggest that A. oryzae mutants lacking α-1,3-glucan in their cell wall can be cultured at a high cell-density. The rate of glucose consumption and pH of the culture broth were comparable for the tripleΔ-cutL1 and wild-type-cutL1 strains (Fig. (D) and (E)).
Fig. 4. CutL1 productivity of Aspergillus oryzae wild-type-cutL1 and tripleΔ-cutL1 strains grown in the presence of 6% glucose.
Discussion
Filamentous fungi such as aspergilli are used in industrial fermentation processes to produce commercially valuable enzymes and low molecular weight compounds such as organic acids and drug leads.Citation30) However, under liquid culture conditions filamentous fungi form hyphal pellets, which lowers cell density and prevents high fermentation productivity. A. nidulans mutants in which the agsB gene encoding the major α-1,3-glucan synthase was disrupted (agsBΔ) lost most of the α-1,3-glucan from their cell wall and formed dispersed hyphae in liquid culture rather than pelletsCitation4); we hypothesized that the same might be true in the industrial fungus A. oryzae. We disrupted the three genes encoding α-1,3-glucan synthases in A. oryzae (agsAΔagsBΔagsCΔ), and we found that the hyphae of the tripleΔ strain lost most of α-1,3-glucan in the cell wall (Fig. (C)) but still formed small pellets under liquid culture conditions. The pellets formed by the tripleΔ strain were smaller than those formed by the wild-type strain under liquid culture conditions (Fig. (A) and (B)).
Whereas the wild-type strain of the human pathogen A. fumigatus, which possesses three α-1,3-glucan synthase genes, forms aggregated, germinating conidia, the mutant in which three α-1,3-glucan synthase genes are disrupted does not.Citation31,32) Furthermore, in the industrial fungus Aspergillus niger, culture conditions such as glucose concentration, temperature, pH, and ionic strength of the culture broth have been shown to affect the aggregation of conidia and hyphae.Citation33,34) Also in A. niger, melanin located on the surface of conidia contribute to the aggregation of conidia into pellets.Citation35) It is noteworthy that an A. nidulans mutant lacking melanin did not produce cell-wall α-1,3-glucan.Citation36) Together, these data suggest that melanin, in combination with α-1,3-glucan, is involved in the aggregation of conidia and hyphae. However, in the present study, A. oryzae wild-type did not produce melanin under liquid culture conditions and yet still formed aggregated hyphal pellets (Figs. ).
Contrary to the completely dispersed hyphae of A. nidulans agsBΔ and agsAΔagsBΔ,Citation4) the small hyphal pellets produced by A. oryzae tripleΔ suggest that other genes are involved in hyphal aggregation besides the three ags genes. He et al. recently reported that amyD and amyG are involved both in α-1,3-glucan biosynthesis and hyphal aggregation in A. nidulans.Citation18) Therefore, we hypothesized that orthologs of amyD and amyG may be involved in hyphal aggregation in A. oryzae tripleΔ. We disrupted the amyD and amyG genes in the A. oryzae tripleΔ strain to construct a quintupleΔ strain; however, A. oryzae quintupleΔ still formed small hyphal pellets, suggesting that neither AmyD nor AmyG was involved in hyphal aggregation in A. oryzae tripleΔ. Analysis of the composition of the hyphal cell wall showed that most of the α-1,3-glucan was lost in A. oryzae tripleΔ and quintupleΔ (Fig. (C)). Therefore, besides α-1,3-glucan, other cell-wall components must be responsible for the remaining hyphal aggregation in A. oryzae tripleΔ under liquid culture conditions. We are now searching for these additional factors.
Although we did not achieve complete hyphal dispersion in A. oryzae under liquid culture conditions, the tripleΔ and quintupleΔ strains of A. oryzae did form smaller hyphal pellets than did the wild-type strain. We hypothesized that the larger surface area provided by the smaller pellets formed by A. oryzae tripleΔ and quintupleΔ might result in improved fermentation productivity. A. oryzae tripleΔ and quintupleΔ, whether or not also overexpressing CutL1, formed hyphal pellets that were approximately 30% smaller than those of the wild-type strain when cultured in YPD/YPDS (Figs. and ) and YPM (Fig. ) liquid media. Furthermore, the pH of the culture broth in which tripleΔ or quintupleΔ strain was grown was slightly higher than that of the culture broth in which the wild-type strain was grown at 24–48 h of culture. Together, these results suggest that the small mycelial pellets of the tripleΔ and quintupleΔ strains, whether or not also overexpressing CutL1, meant that fewer cells inside the pellets were subject to anaerobic conditions, leading to an increase in the total energy yield by respiration and consequently an increase in biomass (Figs. (B) and (C)). In addition, the sizes of the mycelial pellets produced by the wild-type strain were larger than those produced by the tripleΔ and quintupleΔ strains (Figs. (B) and (B)). Because the cells inside the pellets are considered to be under anaerobic conditions and therefore cannot fully metabolize sugars via the tricarboxylic cycle, the cells inside the larger pellets of the wild-type strain are thought to produce organic acids. The total number of cells under anaerobic conditions in the hyphal pellets of the wild-type strain was considerably larger than those in the hyphal pellets of the tripleΔ and quintupleΔ strains, but the pH of the culture broth in which the wild-type strain was grown was only slightly lower than that of the culture broth in which the tripleΔ and quintupleΔ strains were grown. We therefore hypothesized that the increased biomass of the tripleΔ and quintupleΔ strains under liquid culture conditions may improve fermentation productivity.
Because the A. oryzae secretory enzyme CutL1 is used to degrade polyesters such as polybutylene succinate-co-adipate,Citation19) we selected CutL1 as a model enzyme for comparing enzyme productivity among the A. oryzae wild-type, tripleΔ, and quintupleΔ strains. In YPM medium, tripleΔ-cutL1 and quintupleΔ-cutL1 strains formed smaller hyphal pellets than wild-type-cutL1 (Fig. (B)). As expected, the CutL1 productivity of the tripleΔ-cutL1 and quintupleΔ-cutL1 strains were significantly higher than those of the wild-type strain (Fig. (E)). Based on the data presented in Fig. (B) and (C), no significant differences were observed in CutL1 secretion among the wild-type-cutL1 (37.6 ± 0.6 ng CutL1/mg mycelial dry weight), tripleΔ-cutL1 (44.6 ± 2.8 ng/mg), and quintupleΔ-cutL1 (43.9 ± 5.8 ng/mg) strains. This suggests that the increase in the production of CutL1 per batch in the A. oryzae tripleΔ strain was attributable to the increased biomass of tripleΔ-cutL1, and therefore this strain may be highly beneficial for use in industrial fermentation processes.
In the tripleΔ and quintupleΔ strains, the production of endogenous amylase (AmyB) was also increased compared to that in the wild-type strain (Fig. (D)), suggesting that the protein secretion machinery in the α-1,3-glucan-deficient mutants remains genetically unaltered. This result suggests that α-1,3-glucan-deficient mutants may be used to produce not only CutL1 but also other secretory proteins under liquid culture conditions.
Small hyphal pellets similar to those produced by the tripleΔ strain can be achieved by increasing the inoculum size of wild-type conidia (1 × 105/mL), which leads to an improvement in CutL1 productivity (Fig. S5). Therefore, regardless of the presence or absence of α-1,3-glucan in the cell wall, when small pellets can be formed, enzyme productivity can be improved. However, it is not feasible to prepare and inoculate a larger number of conidia (e.g. 1 × 107/mL conidia) on an industrial scale. Since higher productivity was achieved by the tripleΔ strain even with a small inoculum size (e.g. 1 × 105/mL conidia) (Fig. S5), α-1,3-glucan-deficient strains are still likely to have beneficial industrial uses.
Cheap carbon sources such as sugar cane and corn steep liquor are commonly used in industrial fermentation processes. Such carbon sources generally contain high concentrations of sucrose, glucose, fructose, or glucose oligomers, which induce carbon catabolite repression and limit fermentation productivity. Therefore, we examined the fermentation productivity of A. oryzae wild-type-cutL1 and tripleΔ-cutL1 strains in the presence of a high concentration of glucose (6% w/v) (Fig. (A) and (B)). Interestingly, the tripleΔ-cutL1 strain produced a significantly higher amount of CutL1 (Fig. (B)) and a greater amount of biomass at 48 h of culture compared with the wild-type-cutL1 strain (Fig. (C)). After 48 h of culture, the growth rate of the wild-type strain was decreased, but that of the tripleΔ-cutL1 strain was not. This decrease in the growth rate of the wild-type strain might be due to the formation of larger pellets, leading to an increase in the number of cells under anaerobic conditions inside the pellets and to the observed decrease in protein productivity after 48 h of culture. In contrast, the cells of the tripleΔ-cutL1 strain were kept under aerobic conditions until the end of the culture period and a higher protein productivity compared with the wild-type strain was observed. Consequently, the constructed A. oryzae tripleΔ strain may be useful for improving the productivity of industrial fermentation processes.
In the present study, we succeeded in improving the enzyme productivity of the industrial fungus A. oryzae by constructing mutants lacking α-1,3-glucan in their cell wall. Removal of α-1,3-glucan from the cell wall of A. oryzae hyphal cells led to the formation of smaller hyphal pellets. The smaller hyphal pellets formed by the A. oryzae tripleΔ strain meant that more hyphal cells were subject to aerobic conditions, which increased the efficiency of the respiratory energy generation that is required for enzyme production. Contrary to the complete hyphal dispersion of the A. nidulans agsBΔ and agsAΔagsBΔ mutants developed previously, the A. oryzae tripleΔ and quintupleΔ strains still formed small hyphal pellets, suggesting that other factors affect hyphal aggregation in A. oryzae. Our group is now examining the remaining factors that affect hyphal aggregation, and complete hyphal dispersion in A. oryzae would lead to better fermentation productivity. Our approach should be applicable to other industrial fungi that have α-1,3-glucan in their cell walls.
Author contributions
KM, AY, and KA conceived and designed the experiments. KM, ZS, MS, and KG carried out the construction of the fungal mutants. KM performed the essential experiments and analyzed the data. KM, AY, and KA wrote the paper. All authors discussed the results and commented on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Bio-oriented Technology Research Advancement Institution (BRAIN) and the Japan Society for the Promotion of Science under Grant-in-Aid for Scientific Research (B) [26292037].
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2016.1209968.
TBBB_1209968_Supplementary_Tables.docx
Download MS Word (18.7 KB)TBBB_1209968_Supplementary_Figures.ppt
Download MS Power Point (1.5 MB)TBBB_1209968_Supplementary_Figure_Legends.docx
Download MS Word (21.1 KB)References
- Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of koji mold and exploration of its future. DNA Res. 2008;15:173–183.10.1093/dnares/dsn020
- Kobayashi T, Abe K, Asai K, et al. Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007;71:646–670.10.1271/bbb.60550
- Tamano K. Enhancing microbial metabolite and enzyme production: current strategies and challenges. Front. Microbiol. 2014;5:718-1–718-5.
- Yoshimi A, Sano M, Inaba A, et al. Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS ONE. 2013;8:e54893.10.1371/journal.pone.0054893
- Beauvais A, Fontaine T. Aspergillus cell wall and biofilm. Mycopathologia. 2014;178:371–377.10.1007/s11046-014-9766-0
- Hagiwara D, Yoshimi A, Sakamoto K, et al. Response and adaptation to cell wall stress and osmotic stress in Aspergillus species. In: Takagi H, Kitagaki H, editors. Stress biology of yeasts and fungi, Chapter 13. Tokyo: Springer; 2015. p. 199–218.
- Fontaine T, Simenel C, Dubreucq G, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000;275:27594–27607.
- Bernard M, Latgé JP. Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 2001;39:9–17.10.1080/744118873
- Mizutani O, Shiina M, Yoshimi A, et al. Substantial decrease in cell wall α-1,3-glucan caused by disruption of the kexB gene encoding a subtilisin-like processing protease in Aspergillus oryzae. Biosci. Biotech. Biochem. 2016. Published online: 15 Mar 2016. doi:10.1080/09168451.2016.1158632
- Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 2004;53:153–165.10.1111/j.1365-2958.2004.04131.x
- Rappleye CA, Goldman WE. Defining virulence genes in the dimorphic fungi. Annual Rev. Microbiol. 2006;60:281–303.10.1146/annurev.micro.59.030804.121055
- Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. USA. 2007;104:1366–1370.10.1073/pnas.0609848104
- Maubon D, Park S, Tanguy M, et al. AGS3, an α(1–3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Gent. Biol. 2006;43:366–375.10.1016/j.fgb.2006.01.006
- Beauvais A, Bozza S, Kniemeyer O, et al. Deletion of the α-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathogens. 2013;9:e1003716.10.1371/journal.ppat.1003716
- Fujikawa T, Kuga Y, Yano S, et al. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 2009;73:553–570.10.1111/mmi.2009.73.issue-4
- Nishimura M. Cell wall reorganization during infection in fungal plant pathogens. Physiol. Mol. Plant Pathol. 2016;95:14–19. doi:10.1016/j.pmpp.2016.03.005
- Fujioka T, Mizutani O, Furukawa K, et al. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell. 2007;6:1497–1510.10.1128/EC.00281-06
- He X, Li S, Kaminskyj SGW. Characterization of Aspergillus nidulans α-glucan synthesis: roles for two synthases and two amylases. Mol. Microbiol. 2014;91:579–595.10.1111/mmi.2014.91.issue-3
- Maeda H, Yamagata Y, Abe K, et al. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005;67:778–788.10.1007/s00253-004-1853-6
- Mizutani O, Kudo Y, Saito A, et al. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Gent. Biol. 2008;45:878–889.10.1016/j.fgb.2007.12.010
- Kühn R, Torres RM. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 2002;180:175–204.
- Mizutani O, Masaki K, Gomi K, et al. Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl. Environ. Microbiol. 2012;78:4126–4133.10.1128/AEM.00080-12
- Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132.10.1002/(ISSN)1097-0061
- Mizutani O, Nojima A, Yamamoto M, et al. Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae. Eukaryot. Cell. 2004;3:1036–1048.10.1128/EC.3.4.1036-1048.2004
- Takahashi T, Maeda H, Yoneda S, et al. The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol. Microbiol. 2005;57:1780–1796.10.1111/j.1365-2958.2005.04803.x
- Gomi K, Iimura Y, Hara S. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 1987;51:2549–2555.
- DuBois M, Gilles KA, Hamilton JK, et al. A colorimetric method for the determination of sugars. Nature. 1951;168:167.10.1038/168167a0
- DuBois M, Gilles KA, Hamilton JK, et al. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956;28:350–356.10.1021/ac60111a017
- Minetoki T. The expression system of heterologous genes in aspergilli (in Japanese). Kagaku To Seibutsu. 2000;38:831–838.10.1271/kagakutoseibutsu1962.38.831
- Abe K, Gomi K, Hasegawa F, et al. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153.10.1007/s11046-006-0049-2
- Fontaine T, Beauvais A, Loussert C, et al. Cell wall α1-3glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Gent. Biol. 2010;47:707–712.10.1016/j.fgb.2010.04.006
- Henry C, Latgé JP, Beauvais A. α1,3-glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell. 2012;11:26–29.10.1128/EC.05270-11
- Grimm LH, Kelly S, Krull R, et al. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 2005;69:375–384.10.1007/s00253-005-0213-5
- Fleißner A, Dersch P. Expression and export: recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 2010;87:1255–1270.10.1007/s00253-010-2672-6
- Priegnitz BE, Wargenau A, Brandt U, et al. The role of initial spore adhesion in pellet and biofilm formation in Aspergillus niger. Fungal Genet. Biol. 2012;49:30–38.10.1016/j.fgb.2011.12.002
- Polacheck I, Rosenberger RF. Aspergillus nidulans mutant lacking α-(1,3)-glucan, melanin, and cleistothecia. J. Bacteriol. 1977;13:650–656.
