Abstract
Satoshi Ōmura, Professor Emeritus at Kitasato University, was awarded the Nobel Prize for his discovery of a substance of tremendous value to mankind from a microorganism. As a researcher who regularly deals with enzymes produced by microorganisms and a person engaged in microorganism-based business, Professor Ōmura’s Nobel Prize fills me with great pride and joy. It is perhaps not surprising that this Nobel Prize-winning research would emerge from Asia, specifically Japan, where people live in harmony with nature rather than try to conquer it. At Amano Enzyme Inc., we devote ourselves to searching for novel enzymes from microorganisms. While incorporating my own experiences, I will recount the stories of a few discoveries of valuable enzyme-producing microbes in soil and bacterial strain libraries. I will also briefly introduce microbial strain library construction as a tool for facilitating the identification of the desired producing bacteria.
Key words:
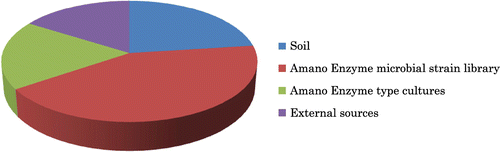
Currently, Amano Enzyme Inc. supplies a variety of enzyme preparations for a wide range of applications, including pharmaceuticals (predominately gastrointestinal agents), dietary supplements, bioconversion of pharmaceutical intermediates, diagnostic reagents, food processing, technical industry, and environmental industry. Most of these enzymes are derived from microorganisms. We work with well over 100 enzyme-producing microorganisms, which were obtained from the following sources: direct isolation from soil and other natural sources (23%), our own company’s microbial strain library (43%), external type culture collections (18%), and other external sources (16%) (Fig. ). Taxonomically, filamentous fungi and bacteria (in nearly equal numbers) account for over 90% of these microorganisms, while the remaining 10% comprises yeasts and basidiomycetes.
In this manuscript, I will introduce three cases of discoveries of microorganisms in Amano Enzyme Inc., two of which were discovered by screening soil samples, and the one in our microbial strain library. For the development of the novel enzymes in the first and second cases, we received Awards for Achievement in Technological Research by the Japan Society for Bioscience, Biotechnology, and Agrochemistry (the first award was a joint award with Ajinomoto Co., Inc.). In the final segment, I will emphasize the importance of Amano Enzyme’s microbial strain library as a source for screening novel enzyme-producing microorganisms, and will introduce some of our endeavors to enrich this resource.
Discovery of a transglutaminase-producing bacteriumCitation1,2)
Transglutaminase is a well-known enzyme used for protein crosslinking in a wide range of processed foods such as meat, fish, dairy products, and wheat products. It is one of the most commonly used enzymes worldwide in quantity as a single enzyme protein.
In the late 1980s, through joint research with Ajinomoto Co., Inc., an actinomycete species referred to as strain S-8112 (later identified as a variant of Streptoverticillium mobaraense and currently reclassified as Streptomyces mobaraensis) was discovered to produce transglutaminase at the research center of Amano Enzyme Inc. (formerly Amano Pharmaceutical Co., Ltd., Kita-Nagoya-shi, Aichi Prefecture).
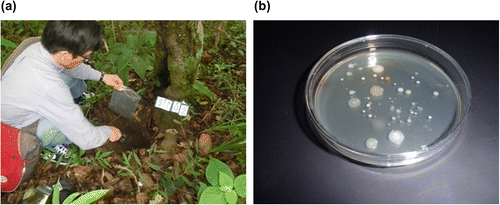
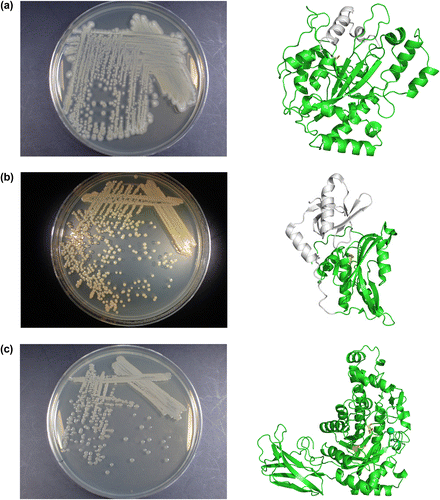
At that time, I was working in a different group at that same research center, but I learned about background of the discovery as follows. In the laboratory, culture medium stocks had been prepared from diverse strains isolated from soil samples for various research purposes. Of these and the numerous culture supernatant samples, about 20 were detected to exhibit activity for production of yellow hydroxamate from N-carboxy-L-glutaminyl-glycine (Cbz-Gln-Gly) and hydroxylamine in a simple method developed to measure animal transglutaminase activity. Furthermore, one of these 20 samples was also discovered to be capable of gelatinizing soy milk. Following this discovery, innovative efforts were made to screen for halo formation due to protein aggregation using a plate with plasma incorporated in the culture medium as a substrate protein; these efforts culminated in the selection of strain S-8112 (Fig. (a)).
Fig. 2. Enzyme-producing bacteria and their respective enzyme conformations.
At that time, transglutaminases were known in some quarters to have potential applications in food processing; however, the only sources of the enzymes were guinea pig livers and other animal organs, thus making practical application difficult. The discovery of a microorganism which enabled unlimited production of transglutaminases by fermentation was a key factor in its subsequent practical application. The achievements of Akira Matsuura and the late Hiroyasu Ando, my colleagues responsible for this discovery, and the great insight of the people at Ajinomoto Co., Inc. who quickly realized the usefulness of transglutaminase, are highly laudable. As a result of these achievements, transglutaminases have dramatically affected the global food industry.
Discovery of a protein-glutaminase-producing bacteriumCitation3,4)
Several years following the discovery of a transglutaminase-producing bacterium, we aimed to supply different novel enzymes that could greatly contribute to society, and thereby commenced a series of investigations on protein-modifying enzymes. Because widely applicable enzymes are often simple, we began our search by simultaneously screening for a list of target enzymes known to catalyze reactions in a living body, such as enzymes which crosslink proteins via oxidation, enzymes which form and cleave disulfide bonds in protein, protein kinases, and protein deamidases. Known reactions in a living body can be considered to be safe for food use rather than reactions never known in living cell. To screen for protein deamidase-producing microoraganisms, the cultural media of soil isolates and of strains from the Amano Enzyme Inc. microbial strain library were screened for the release of ammonia from Cbz-Gln-Gly, the substrate used to measure transglutaminase activity. For the soil samples, we conducted enrichment cultures with Cbz-Gln-Gly as the sole nitrogen source. Through a series of enrichment cultures of 320 soil samples, 446 strains of bacteria and fungi were isolated. We combined these with the 350 strains from the microbial library and measured ammonia release activity in culture medium for 794 strains. Two positive strains were identified; these strains presented with nearly identical visual and cultural characteristics, and the enzymes derived from these strains possessed very similar properties. Therefore, we selected strain No. 9670, which exhibited 20–30% higher activity in culture medium than the other, and proceeded to purify and characterize the enzyme produced by the strain. We determined that this strain extracellularly produced an enzyme that deamidates glutamine residues within proteins like casein and gluten familiar in food industry, converting them to glutamate residues without cleaving peptide bonds. This enzyme had precisely the desired protein-deamidating activity. Strain No. 9670 was identified as a new species of the genus Chryseobacterium and named C. proteolyticum (Fig. (b)).
Incidentally, when I later checked the isolation source of the two positive strains, it was found that they were cultivated from soil samples collected in the same location on the same day. At that time, I was working at Tsukuba research laboratory of Amano Enzyme; I traveled to neighboring farms, livestock barns, factories, and forests to collect soil samples for enzyme screening. One day, when I went to a stream used for irrigation around a company housing unit with my children to catch crayfish, I collected soil samples from a field around the stream and the bank of it; these were the samples that contained the two positive strains.
This deamidating enzyme catalyzes an extremely simple reaction that can dramatically change the physical properties of the substrate protein. Thus, it was anticipated to be useful for a variety of applications, and they were subsequently developed. Furthermore, no homologous protein in both a primary structure and a conformation to this enzyme has yet to be discovered. As having a complete novelty, it was named protein-glutaminase. In addition, through a similar series of screening studies, we discovered oxidative crosslinking of proteins by laccase and other multi-copper oxidases.Citation5)
Discovery of a β-amylase-producing bacteriumCitation6)
The two enzymes discussed thus far were derived from microorganisms discovered in soil. In contrast, the enzyme discussed next was derived from a microorganism found not in soil, but in the Amano Enzyme microbial strain library.
β-Amylase is widely used to produce maltose from starch. Maltose is produced in large quantities as the raw material for maltitol production. Due to its milder sweetness than sucrose, maltose is also used to sweeten Japanese confections as the form of maltose syrup and crystalline maltose. As a uniquely Japanese application, β-amylase has been used for many years to preserve the softness of daifuku-mochi and dango, traditional Japanese confections made from rice. However, when we began searching for a β-amylase, the only enzymes on the market were from plants such as soybean and barley. Although the existence of microbial β-amylase had been known since the 1970s, it had not been industrialized, likely due to barley-derived enzymes being inexpensive and soybean-derived enzymes having relatively high thermostability, which is necessary in some applications. However, after the U.S. announced a policy related to bioethanol several years ago, grain prices soared, and the supply of soybean enzymes dwindled. Furthermore, grain consumption continued to rise in rapidly growing countries such as China and India, leading to great concern over the stability of plant-based enzyme supplies. There were also concerns about the allergy-inducing potential of plant-derived proteins, and allergen labeling is required for plant-derived enzyme preparations used in food processing. Therefore, several years ago, my colleagues and I set about developing and screening for microbial β-amylase, an enzyme that can be supplied stably and does not require allergen labeling.
First, we turned to the Amano Enzyme microbial strain library, as a handful of strains collected and listed as capable of generating β-amylase-like activity by our company in the 1980s. We ultimately discovered a β-amylase-producing bacterium with a thermostability comparable to that of soybean enzymes. This bacterium, one of the previously mentioned handful of strains, had been preserved as Bacillus sp. APC9451, but was later identified as a strain of B. flexus (Fig. (c)).
Enrichment of the Amano microbial strain library
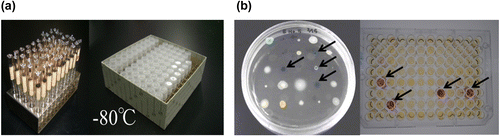
The last example demonstrates that a microbial strain library that includes previously examined strains with the data from research and analysis of those strains can be a powerful tool for screening. At Amano Enzyme, we have worked hard for many years to enrich our microbial strain library, which currently contains a grand total of 13,000 strains. A certain part of these strains are accompanying the results from screenings, and from research and development endeavors undertaken by predecessor people over the course of our company’s long history. The times at which enzymes and other valuable substances are discovered often does not match the times at which society requires those substances. This is especially true in corporate research and development; even if a product is believed to have been developed through scrupulous marketing, opportunities are often missed due to premature emergence of the product or because of competition. When advancements and changes in society result in needs, it is important to quickly supply seeds that meet those needs. Thus, a microbial strain library containing past data is a valuable resource for rapidly discovering microorganisms capable of producing target enzymes (Fig. ).
Fig. 3. Amano Enzyme microbial strain library ((a) left: lyophilized ampule, right: glycerol stock) and an example of screening for enzyme production. ((b) left: plate assay, right: enzyme assay). Arrows (←) indicate positive strains.
In some cases, the microorganisms targeted in the screening are narrowed to specific strains and taxonomic groups in advance. However, due to advances in taxonomy, many old preserved strains have been reclassified. Hence, at Amano Enzyme, we are performing rRNA sequence analyses to re-identify and reclassify the vast majority of our strains.
In recent years, Amano Enzyme has been participating in a project undertaken by the National Institute of Technology and Evaluation (NITE), which has allowed us to add isolates from Vietnam, Mongolia, and Myanmar to our microbial strain library. Although NITE began the Microbial Resources Joint Search Project in Indonesia in 2003, there remained a disparity in the distribution of profits between the counties supplying the gene resources and the countries using them. However, this issue was resolved by the Nagoya Protocol, which was adopted at the 2010 10th Conference of the Parties (COP10). Currently, NITE is encouraging many Japanese corporations to participate in their project, expanding the microbial search project in Asian countries, and constructing a system that allows each participating company to search for and isolate microorganisms to meet their needs. Since joining this project in 2012, Amano Enzyme has added 438 strains from Mongolia, 628 strains from Vietnam, and 578 strains from Myanmar to its library. In Mongolia, we isolated microorganisms from fermented dairy products and soil, focusing on the differences in climate and vegetation. In Vietnam and Myanmar, we collected soil samples from the northern forests and from the southern jungles near the equator; we also collected samples of fish sauce and other fermented foods (Fig. ).
Concluding remarks
When Prof. Ōmura was awarded the Nobel Prize, he modestly commented, “I merely borrowed the power of microbes.” However, behind his discovery of a microorganism and substance with tremendous value to society lies a rare combination of abilities—assiduous effort, steadfast determination, attentiveness and concentration which do not overlook a single phenomenon, unique ideas, a broad perspective, and a personality that attracts others. I imagine that I am far from the only one who feels this way. As a scientist who deals with microorganisms—although for enzymes and not antibiotics—and as a person involved in the fermentation industry on which Japan prides itself, this award was very encouraging.
In this manuscript, I have described a few examples of searches for enzyme-producing microbes undertaken by the enzyme manufacturer Amano Enzyme. In recent years, the desire for enzymes with new functions has led to a boom in protein engineering methods. However, as I mentioned in the beginning, Japanese in particular do not seek to conquer nature, but to live in harmony with it. Screening from nature rather than employing protein engineering methods may very well be the embodiment of this philosophy. I hope all of you will find this beneficial.
Disclosure statement
No potential conflict of interest was reported by the author.
Notes
† This work is a translation of an original work in Japanese in the Japan Society for Bioscience, Biotechnology, and Agrochemistry http://doi.10.1271/kagakutoseibutsu.54.61.
References
- Ando H, Adachi M, Umeda K, et al. Agric Biol Chem. 1989;53:2613.
- Washizu K, Ando K, Koikeda S, et al. Biosci Biotechnol Biochem. 1994;58:82.10.1271/bbb.58.82
- Yamaguchi S, Yokoe M. Appl Environ Microbiol. 2000;66:3337.10.1128/AEM.66.8.3337-3343.2000
- Yamaguchi S, Jeenes DJ, Archer DB. Eur J Biochem. 2001;268:1410.10.1046/j.1432-1327.2001.02019.x
- Yamaguchi S. Japanese patent No. 4137224. 1998. (in Japanese).
- Sugita A, Okada M, Tani A, et al. Bull Appl Glycosci. 2011;1:194.