Abstract
Transglutaminase is an enzyme family responsible for post-translational modification such as protein cross-linking and the attachment of primary amine and/or deamidation of glutamine-residue in proteins. Medaka (Oryzias latipes), a recently established model fish, has similar functional proteins to those characterized in mammals. Previously, we found the apparent orthologues that correspond to human transglutaminases in medaka. In this study, regarding the medaka orthologue of human tissue-type transglutaminase (OlTGT), recombinant protein was expressed in an active form in bacteria cultured at low temperature. Using the recombinant protein, we biochemically characterized the enzymatic activity and also obtained a monoclonal antibody that specifically recognized OlTGT. Immunochemical analysis revealed that OlTGT was not expressed ubiquitously, unlike its mammalian orthologue, but in primarily limited tissues such as the eye, brain, spinal cord, and gas gland.
Key words:
Transglutaminases (TGases) are a family of Ca2+-dependent enzymes that catalyze protein cross-linking reactions resulting in functional and structural modification in a variety of biological processes.Citation1–3) In mammals, the TGase family consists of eight members (TG1–TG7 and Factor XIII (FXIII)) as isozymes that are characterized by their various distribution and regulatory mechanism of activity. Among the family members, FXIII is involved in blood coagulation by polymerizing of fibrin.Citation4) TG1, TG3 and TG5 have functional roles in skin barrier formation by cross-linking of various structural proteins in differentiating keratinocytes.Citation5,Citation6) TG2, major isozyme, is ubiquitously expressed in mammals and has multiple roles, such as in the stabilization of extracellular matrix proteins, inactivation of transcription factors, and modification of signal transduction molecules.Citation7,Citation8) Furthermore, in the case of TG2, non-catalytic functions have been found, such as GTP binding proteins and interacting proteins for scaffolding. Several related diseases affected by aberrant regulation of TG2 enzymatic activity have been suggested, such as celiac disease, cancer, and neurodegenerative diseases. However, the physiological functions of TG2 have yet to be clarified.
Possible orthologues for TG2 are also found in a wide variety of species including both vertebrates and invertebrates.Citation1) In animals other than mammals, TG2 primarily appears to be required for recovery from injury or barrier formation. However, in mammals and the other animals, the physiological significance of TG2 and its relevance to diseases are still valuable issues to be addressed. Considering the essential and various roles of TG2, unknown functions of this protein will expand upon the related phenomena of this isozyme.
Medaka, a model fish recently used in parallel to zebrafish (Danio rerio), has several advantages for investigating the functions of genes of interest.Citation9–11) Taking advantages of feasible maintenance, high fertility, and short generation period, several kinds of gene-modified fishes have been established and used for drug screening in research areas, including pharmaceutical investigations.Citation12,13) We recently discovered the orthologues of TGases in medaka, which consists of seven genes: OlTGK1, OlTGK2, OlTGK3 (for hTG1), OlTGB (for hFXIII), OlTGT (for hTG2), and OlTGF and OlTGO that do not correspond with specific mammalian TGase.Citation14) In our previous study, biochemical characterization of the OlTGK1 gene product indicates that this enzyme has cross-linking activity and was expectedly localized to the skin epidermis and spine bone. However, for the other TG isozymes in medaka, investigations are ongoing to determine their expression pattern and biochemical data. In the present study, for OlTGT, we carried out these experiments using recombinant proteins and thus obtained monoclonal antibody which is essential for expression analyses of isozymes. Since TG2 is a major isozyme and has multiple functions in mammals, we believe the investigation of OlTGT, as medaka orthologue, including its biochemical properties and expression pattern will contribute to improved understanding of the physiological significance.
Materials and methods
Animal experiments
For use of rat and medaka in a series of experiments, animal care and experiments were carried out according to the Regulations for Animal Experiments in Nagoya University.
Expression and purification of recombinant OlTGT
The plasmid vector harboring OlTGT was obtained from NBRP (National BioResourses Project of Japan, Okazaki, Japan), as clone ID: olsp62a16. The cDNA sequence was analyzed and the data were deposited to DDBJ (LC068826).
The purification of recombinant OlTGT was carried out as previously reported.Citation14) Briefly, using the plasmid vector DNA (olsp62a16) as a template, the possible full-length OlTGT cDNA was amplified by PCR with specific primers, to attach the SmaI and XhoI recognition sites at 5′ and 3′, respectively (forward; 5′-atcccggggctcaatctgtggacattgaac-3, reverse; 5′-atctcgagttactttccaatgatgacgtttc-3′). The amplified cDNA was confirmed its sequence and the SmaI-XhoI fragment was inserted into the expression vector, pET24dHis which includes the initiation codon and enables to express the protein attached with hexahistidine at the N-terminus. E. coli. BL21(DE3)LysS was transformed with the constructed plasmid vector and the transformant was grown in L-broth. Induction of recombinant OlTGT expression was carried out by a standard method using isopropyl β-D-1-thiogalactopyranoside (IPTG).
The harvested bacteria were lysed in lysis buffer (10 mM Tris-Cl, pH 8.0, 10 mM NaCl, 1 mM β-mercaptoethanol, phehylmethylsulfonyl fluoride, and benzamidine) following sonication. The supernatant obtained by centrifugation was applied to TALON (Clontech-TAKARA, Kyoto Japan) and then eluted by elution buffer (wash buffer containing 100 mM imidazole). Then the eluted proteins were subjected to size separation gel chromatography (Superdex-200 increase; GE Healthcare Bio-Sciences AB Uppsala, Sweden).
Measurement of in vitro activity of transglutaminase
The enzymatic activity was measured by incorporation of 5-(biotinamido)pentylamine (bio-Cd, Pierce, IL, USA) into β-casein fixed on the 96 well microtiterplates in a buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 15 mM CaCl2, 1 mM dithiothreitol. After incubation, microtiter well was washed with TBS buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl) containing 0.1% tween-20. After adding strpetavidin-peroxidase (Rockland Immunochemicals Inc., Gilbertsville, PA, USA) and washing, the amounts of the reaction product was measured by color development of the peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB, Sigma-Aldrich, St. Louis, MO).
TG2 enzymatic activity was reported to be inhibited by GTP.Citation15) To examine the inhibitory effect on the enzymatic activity by GTP, the recombinant proteins for human TG2 (Zedira, Darmstadt, Germany) and OlTGT were used for the enzymatic reaction. GTP solution (Sigma-Aldrich), dissolved in 0.1 M Tris-Cl (pH. 8.0), was contained at the indicated concentrations in the reaction as described above.
Establishment of rat hybridoma secreting monoclonal antibody against OlTGT
The purified recombinant proteins for OlTGT was used for immunization to Wister rat as antigen with Freund complete (once) or incomplete (twice) adjuvant in two months at the footpad. The lymphocytes from the immunized rat was prepared and fused with proliferating myeloma (Sp2/0-Ag14) using polyethylenglycol 4000 by a standard method. The hybridomas were selected in HAT medium, and then the supernatants of the grown colonies were analyzed for the production of target antibody by ELISA method. The hybridomas were amplified, cloned, and then grown for production of the antibody against OlTGT (T9D). From the supernatant of cultured cells in the serum-free medium (Hybridoma-SFM, Gibco, Lifetechnologies) monoclonal antibody was affinity-purified using Protein-A immobilized gel by a standard method.
Immunoblotting and immunostaining
The paraffin section of medaka (cab, adult fish) was prepared by standard method after fixation of Davidson solution (acetic acid, formalin and ethanol) for 7 days. The 10 μm section on the slide glass was prepared and de-paraffin was carried out using xylene and ethanol. To the section that was blocked by blocking solution (Vectastain blocking solution including rabbit normal serum, Vector Laboratories; Burlingame, CA, USA), the monoclonal antibody (T9D) diluted with 0.1% BSA-containing PBS was added and then incubated at 4 °C overnight. After washing the section with the wash buffer, in order to amplify the immunoreaction, biotin-labeled secondary antibody was reacted. Then, followed by adding avidin and biotin-peroxidase, color development was performed using 3,3′-diaminobenzidine (DAB) staining. Rat immunoglobulin G was used for negative control. The section was observed under the fluorescence microscope (BZ-9000; Keyence, Osaka, Japan).
Results
Deduced primary structure of OlTGT
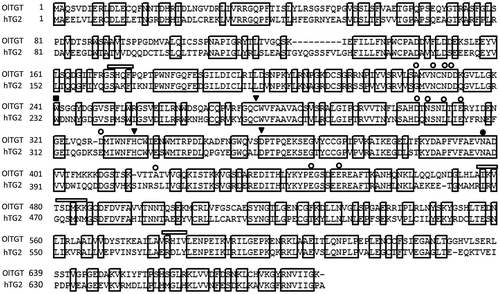
Construction of the phylogenic tree including OlTGT and all the human isozymes let us confirm the cDNA sequence as the orthologue to hTG2 (supplemental figure 1 and Ref. 14). Then, sequence alignment of OTGT with human TG2 is shown in the Fig. . Based on the homology with human TG2, this orthologue consists of 677 putative amino acid residues with overall homology to that of TG2, suggesting that OlTGT contains also four domains: β-sandwich, catalytic core, β-barrels 1 and 2. In addition to the catalytic triad such as Asn, His, and Cys (the active-site residue), the surrounding amino acid residues around the active site (Cys) required for substrate recognition and essential Trp were also preserved in the OlTGT sequence. Moreover, possible calcium binding sites were observed at certain residues with a similar pattern to human TG2. As for GTP binding region, however, there were less homologous sequences, suggesting that OlTGT plays a role only in transamidation activity.
Fig. 1. Sequence alignment of OlTGT and human tissue-type transglutaminase (TG2).
Production and biochemical characterization of recombinant OlTGT in bacteria
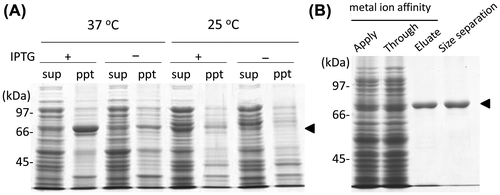
For expression of the recombinant protein, we compared the pattern in two different temperatures for cultivation at 37 or 25 °C. It has previously been reported that OlTGT was harvested as the soluble fraction as expected size of 75 kDa when incubated at the lower temperature (Fig. (A)).Citation14) Under this culture condition, the protein expressed in E. coli was successfully purified to homogeneity by metal affinity-purification, followed by size exclusion chromatography (Fig. (B)). The amounts of purified protein enabled us to perform biochemical characterization studies and to also obtain monoclonal antibodies as an antigen.
Fig. 2. Expression and purification of recombinant OlTGT.
For evaluation of the enzymatic activity, biotin-labeled pentylamine (bio-Cd), as a glutamine-acceptor substrate, was incorporated into the microtiter-well coated β-casein, as a glutamine-donor substrate. Depending on the concentration of this glutamine-acceptor substrate, incorporation of bio-Cd into β-casein was increased in the presence of OlTGT (Fig. (A)). This reaction is also time-dependent (Fig. (B)) and the co-incubation of EDTA in both reactions abrogated the incorporation indicating the enzyme reaction is calcium ion-dependent.
Fig. 3. Enzymatic activity of recombinant OlTGT.
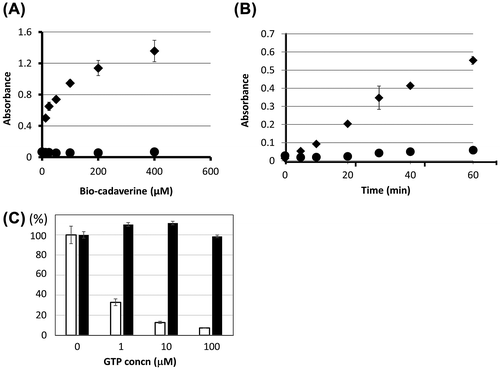
In mammalian TG2, the enzymatic activity is regulated by GTP. Then, we examined its inhibitory effect on the enzymatic activities using recombinant proteins for both hTG2 and OlTGT. As shown in Fig. (C), whereas the activity of hTG2 was inhibited by the presence of GTP in a concentration-dependent manner (1–100 μM), OlTGT did not show any inhibitory regulation. This result is consistent with the fact that there was no reported GTP-interacting sites in the primary sequence of OlTGT.
Immunochemical analyses of OlTGT
A hybridoma that secreted a monoclonal antibody against OlTGT was established using the purified recombinant protein as the antigen. Upon analysis, since confirmation of the cross-reactivity against other medaka TGase isozymes is important, immunoblotting was carried out using each recombinant protein prior to immunohistochemical experiments (supplemental figure 2). The antibody recognized and mainly reacted to OlTGT but slightly cross-reacted with OlTGK1.
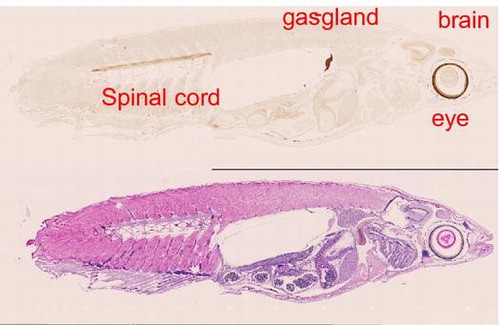
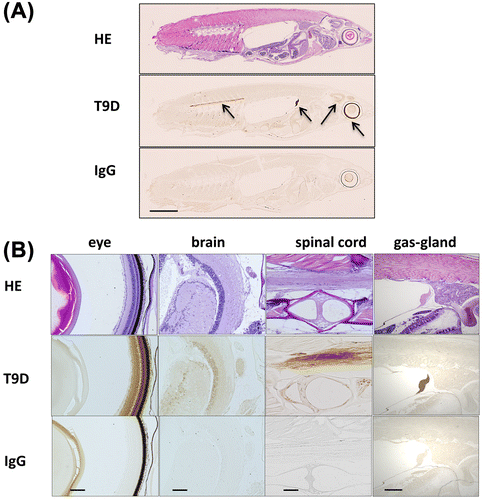
Regarding the immunohistochemical analysis, expression was unexpectedly not ubiquitous as reported in mammals. Higher level of the expression was apparently observed at the site of the eye, brain, spinal cord, and gas gland as shown in Fig. (A). In Fig. (B), the enlarged pictures indicate that in the eye, brain, and spinal cord, staining was specified around the retina, optic tectum, and myelin sheath, respectively. Upon immunostaining, we also confirmed less cross-reactivity using the serial section for OlTGK1 antibody reaction that gave no signal (supplemental figures 3 and 4).
Fig. 4. Immunohistochemical analysis on whole medaka section.
Discussion
Calcium ion-dependent cross-linking reactions catalyzed by TGase are characteristic to vertebrate TGases but also observed in a wide range of the invertebrates TGases. So far, TGases with mammalian-like structures have also been characterized in various organisms including horseshoe crab, Drosophila, grasshopper, and slime mold.Citation1) Among eight human TG isozymes, since tissue-type TGase (TG2) is ubiquitously expressed in and plays multiple roles, a lot of studies on the biochemical properties and physiological functions of this isozyme have been conducted including on other animals. Fish is evolutionally the lowest vertebrate but they maintain several physiological functions of those preserved in mammals. As a well-known model fish, zebrafish has been used in studies of various biological phenomena including TGase.Citation16,Citation17) In addition, in situ expression pattern of one orthologue of TGase in medaka (identical to OlTGB; orthologue for mammalian FXIII) was reported during embryogenesis.Citation18) However, in these studies, investigations at the protein and histological analyses on this isozyme (tissue-type TG; TG2) as TGase family have not been included.
In this fish model, medaka, we very recently identified all the orthologues of human TGases some of which apparently corresponded with the eight mammalian isozymes in medaka genes.Citation14) OlTGT (T means tissue-type), obviously the orthologue for mammalian TG2, has a total deduced molecular mass of 75 kDa, which is similar to human TG2 obtained from the results of recombinant proteins (Fig. ). Although we did not confirm with the other possible translation initiation site in the 5′ flanking region, it would be greatly possible our examined cDNA sequence encoded open reading frame because of high homology with human TG2 sequence. As shown in Fig. , when compared to human TG2, residues essential for the calcium-dependent cross-linking reaction were mostly conserved, which is consistent with the biochemical data.
To characterize the biochemical properties of OlTGT, we produced recombinant protein in bacteria, which was successfully expressed at the lower temperature probably due to sterically stabilized form. These experiments demonstrated that the enzyme was sterically reproducible and active during bacterial expression. Using this sterically stabilized protein as an antigen, we obtained biochemical data and a monoclonal antibody used for the tissue distribution analysis. As expected, the purified recombinant protein for OlTGT showed the apparent enzymatic activity in a calcium-ion dependent manner. Only the difference in the enzymatic property is that OlTGT was not affected by GTP, which inhibits the enzymatic activity of transamidation of mammalian TG2.Citation1–3,15) This result was expected because there is less homologous on the GTP-binding region between human TG2 and OlTGT (Fig. ) This tendency is also found in the case of frog TG2 orthologue (Xenopus laevies; NP_001085410). It is interesting that medaka and frog orthologues for TG2 do not have such a GTP-binding region from the aspect of molecular evolution, but its physiological significance is currently unknown.
The immunohistochemical analysis indicated the data that OTGT is expressed in limited tissues: eye, brain, spinal cord, and gas gland. It is notable that OlTGT is expressed not ubiquitously, which is different property from mammalian TG2.
In the eye, the expressed areas are concentrated around the retina, which was observed in another fish (goldfish) where TG activity is essential for recovery from injury.Citation19) The precise significance of mammalian TG2 in brain remains unknown, however, evidence suggests that it may be involved in IL-2 cross-linking and overexpression may lead some cytotoxic factors to oligodendrocyte.Citation20) As for spinal cord, spontaneous recovery from injury was investigated in adult zebrafish.Citation21) OlTGT may be involved in the event since TG2 contribute to recovery of tissue injury. Although the expressions of gas gland were first found in the fish, identification of substrates might give a clue to know the physiological significance.
In conclusion, we characterized OlTGT, an orthologue for mammalian TG2 involved in various biological processes, both biochemically and immunohistochemically. Limited expression rather than ubiquitous distribution was observed in the eye, brain, spinal cord and gas gland in this organism, which has a possibility for novel functions rather than mammals. In addition to analysis on substrates, the functional significance of this expression will be clarified by tissue-specific deletion, which is possible by using a genome editing system. Research is ongoing in these areas.
Author contributions
The first (Y.T.) and the second (Y.W.) authors performed the experiments with equal contribution. The other authors (K.O., H.T., H.H.) support the experiments and data analyses. The last author (K.H.) organized the planning and completion of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by Scientific Research (B) [grant number 26292192] and Young Scientists Research [grant number 26860500].
Supplemental material
Supplemental material for this article can be accessed at http://dx.doi.org/10.1080/09168451.2016.1256757.
TBBB_1256757_Supplemental_Material.zip
Download Zip (5.6 MB)Acknowledgments
The plasmid vector encoding OlTGT cDNA was provided by National BioResourses Project of Japan, Okazaki, Japan. The technical assistance of medaka tissue sections were kindly given by Mr. T. Nishimaki and Dr. H. Oota (Kitasato Univ.).
Notes
Abbreviations: bio-Cd, 5-(biotinamido)pentylamine; BSA, bovine serum albumin; DTT, dithiothreitol; IPTG, β-D-1-thiogalactopyranoside; PBS, Phosphate-buffered saline; TBS, Tris-buffered saline; TGase, transglutaminase.
References
- Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156.10.1038/nrm1014
- Eckert R, Kaartinen M, Nurminskaya M, et al. Transglutaminase regulation of cell function. Physiol Rev. 2014;94:383–417.10.1152/physrev.00019.2013
- Hitomi, K, Kojima, S, Fesus, L, editors. Transglutaminases. Multiple functional modifiers and tragets for new drug discovery. Tokyo: Springer; 2015.
- Muszbek L, Bereczky Z, Bagoly Z, et al. Factor XIII: a coagulation factor with multiple plasmatic and cellular functions. Physiol Rev. 2011;91:931–972.
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340.10.1038/nrm1619
- Hitomi K. Transglutaminase in skin epidermis. Eur J Dermatol. 2005;15:313–319.
- Iismaa SE, Mearns BM, Lorand L, et al. Transglutaminases and disease: lessons from genetically engineered mouse models and inherited disorders. Physiol Rev. 2009;89:991–1023.10.1152/physrev.00044.2008
- Kanchan K, Fuxreiter M, Fésüs L. Physiological, pathological, and structural implications of non-enzymatic protein–protein interactions of the multifunctional human transglutaminase 2. Cell Mol Life Sci. 2015;72:3009–3035.10.1007/s00018-015-1909-z
- Lin C-Y, Chiang C-Y, Tsai H-J. Zebrafish and Medaka: new model organism for modern biomedical research. J Biomed Sci. 2016;23:19–30.10.1186/s12929-016-0236-5
- Kinoshita M, Murata K, Naruse K, et al. Medaka: biology, management, and experimental protocols. Ames (IA): Wiley-Blackwell; 2009.
- Kirchmaier S, Naruse K, Wittbrodt J, et al. The genomic and genetic toolbox of the teleost Medaka (Oryzias latipes). Genetics. 2015;199:905–918.10.1534/genetics.114.173849
- Matsui H, Ito H, Taniguchi Y, et al. Proteasome inhibition in medaka brain induces the features of Parkinson’s disease. J Neurochem. 2010;115:178–187.10.1111/j.1471-4159.2010.06918.x
- To TT, Witten PE, Renn J, et al. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development. 2012;139:141–150.10.1242/dev.071035
- Kikuta A, Furukawa E, Ogawa R, et al. Biochemical characterization of Medaka (Oryzias latipes) transglutaminases, OlTGK1 and OlTGK2, as orthologues of human keratinocyte-type transglutaminase. PLoS ONE. 2015;10:e0144194.10.1371/journal.pone.0144194
- Achyuthan KE, Greenberg CS. Identification of a guanosine triphosphate-binding site on guinea pig liver transglutaminase. J Biol Chem. 1987;262:1901–1906.
- Deasey S, Grichenko O, Du S, et al. Characterization of the transglutaminase gene family in zebrafish and in vivo analysis of transglutaminase-dependent bone mineralization. Amino Acids. 2012;42:1065–1075.10.1007/s00726-011-1021-0
- Deasey S, Nurminsky D, Shanmugasundaram S, et al. Transglutaminase 2 as a novel activator of LRP6/β-catenin signaling. Cell Signal. 2013;25:2646–2651.10.1016/j.cellsig.2013.08.016
- Koh D, Inohaya K, Imai Y, et al. The novel medaka transglutaminase gene is expressed in developing yolk veins. Gene Expr Patterns. 2004;4:263–266.10.1016/j.modgep.2003.11.004
- Sugitani K, Ogai K, Hitomi K, et al. A distinct effect of transient and sustained upregulation of cellular factor XIII in the goldfish retina and optic nerve on optic nerve regeneration. Neurochem Intl. 2012;61:423–432.10.1016/j.neuint.2012.06.004
- Eitan S, Schwartz M. A transglutaminase that converts interleukin-2 into a factor cytotoxic to oligodendrocytes. Science. 1993;261:106–108.10.1126/science.8100369
- Vajn K, Suler D, Plunkett JA, et al. Temporal profile of endogenous anatomical repair and functional recovery following spinal cord injury in adult zebrafish. PLoS ONE. 2014;9:e105857.10.1371/journal.pone.0105857
