Abstract
Dopa decarboxylase (DDC) protein is involved in the synthesis of dopamine and serotonin. Here, we show that in the silkworm Bombyx mori, a novel DDC splicing variant is selectively expressed in the brain and subesophageal ganglia. In Drosophila melanogaster, a neuron-specific isoform of DDC is known to be alternatively spliced in a similar manner.
Monoamines act as neurotransmitters and neuromodulators in the nervous systems of invertebrates and vertebrates. In Drosophila melanogaster, monoamines regulate various physiological processes, such as feeding, memory, and sexual behavior.Citation1) Dopamine, a monoamine, is converted from L-3,4-dihydroxyphenylalanine by dopa decarboxylase (DDC). DDC also synthesizes serotonin from 5-hydroxy-L-tryptophan.Citation2) In Bombyx mori, elevation in the dopamine level in the brain–subesophageal ganglia stimulates diapause hormone mRNA expression.Citation3,4) In the insect epidermis, DDC synthesizes dopamine as a reaction intermediate involved in cuticular sclerotization.Citation5–7)
In D. melanogaster, different DDC mRNAs are detected in the epidermis and central nervous system (CNS).Citation8) Tissue-specific DDC mRNAs are generated by alternative splicing from a primary transcript of the DDC gene in Drosophila. The epidermal DDC mRNA contains three exons, whereas the CNS DDC mRNA contains four exons, including an additional exon (Fig. ). The additional exon of CNS mRNA is predicted to add 33–35 amino acids to the N-terminus of the epidermal DDC protein.Citation9) In B. mori, the DDC gene is expressed in the epidermis, testis, ovary, silk gland, midgut, and fat body. The epidermal DDC mRNA contains seven exons and encodes a DDC protein comprising 478 amino acid residues.Citation10) Nevertheless, the structure of the 5′-untranslated region (5′-UTR) remains unknown. Moreover, DDC splicing variants of B. mori CNS have not been reported.
Fig. 1. Structures of the dopa decarboxylase (DDC) alternative splicing isoforms of D. melanogasterCitation9) and B. mori (this study).
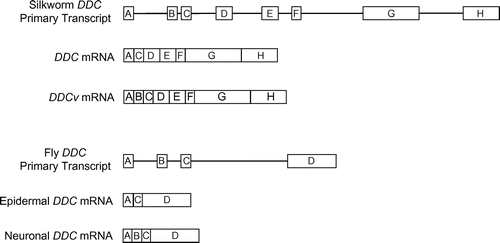
Fig. 2. Sequences of exons A and B of the silkworm (B. mori) DDCv cDNA.

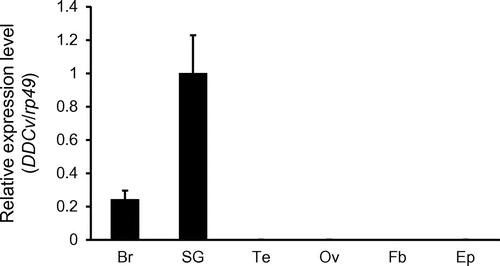
Fig. 3. Tissue-specific expression analysis of dopa decarboxylase variant (DDCv) RNA by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in larvae.
To analyze the 5′-UTR structure of the B. mori DDC mRNA and to facilitate the discovery of a novel DDC splicing variant, a complementary DNA (cDNA) library was prepared from the total RNA isolated from the CNS of B. mori larvae. The 5′-end sequences of DDC mRNAs (5′-DDC) were determined by 5′ rapid amplification of cDNA ends (RACE). We obtained two 5′-DDC species: a novel DDC splicing variant (DDCv) and the DDC mRNA reported by the preceding study.Citation10) The full length of DDCv cDNA was obtained by use of primers designed in exon B and exon H (Fig. ). We identified exon A as the 5′-UTR of DDC mRNA and DDCv mRNA. A previous study reported that DDC mRNA consists of seven exons, namely exon A, C, D, E, F, G, and H.Citation10) Moreover, we clarified that DDCv mRNA consists of eight exons, namely exon A, B, C, D, E, F, G, and H. The predicted initiation codon of DDC mRNA was located in exon C. In contrast, the potential initiation codon of DDCv mRNA was located in exon B, and DDCv mRNA is predicted to encode 490 amino acid residues (Fig. ). Although these N-terminal sequences of the fly and silkworm neuronal DDC isoforms show weak similarity, conserved putative motif could not be found in these sequences.
To investigate tissue-specific expression of the DDCv mRNA, we performed quantitative reverse transcription polymerase chain reaction (qRT-PCR). qRT-PCR analyses showed that DDCv mRNA is highly detected in the brain and SG, but hardly in the other tissues (Fig. ). In particular, the expressional level of DDCv mRNA in the SG was approximately fivefold higher than that in the brain (Fig. ). The expression level of DDC mRNA could not be analysed by qRT-PCR because it is difficult to design primers that selectively amplify the DDC isoform. By RT-PCR and agarose gel electrophoresis, we confirmed that DDC mRNA is expressed in all tissues shown in Fig. (data not shown).
DDCv mRNA was highly expressed in the SG, supporting a report that showed that DDC was detected in only four cells of the SG by in situ hybridization.Citation10) DDCv mRNA was detected in the brain by RT-PCR in the present study, whereas DDC mRNA was not detectable in the brain by in situ hybridization.Citation10) The expression level of DDCv mRNA in the brain might be insufficient for detection by in situ hybridization. Dopamine elevates the level of mRNA for the diapause hormone in Br and SG of the silkworm.Citation3) We reared silkworm larvae of commercial strain under a condition to lay a diapause eggs. Selective expression of DDCv mRNA in the brain and SG suggests that DDCv protein may induce embryonic diapause. In both B. mori and D. melanogaster, alternatively spliced neuronal mRNAs retained additional exons that may encode the short amino acid sequence to the hypodermal DDC proteins. Although the physiological roles of the additional exons remain unclear, the evolutionarily conserved mechanism may reflect important regulations of DDC expressions or functions.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2016.1258987.
Disclosure statement
No potential conflict of interest was reported by the authors.
TBBB_1258987_Supplementary_Material.docx
Download MS Word (20.7 KB)References
- Vömel M, Wegener C. Neuroarchitecture of aminergic systems in the larval ventral ganglion of Drosophila melanogaster. PLoS ONE. 2008;3:e1848.10.1371/journal.pone.0001848
- Wright TRF. Phenotypic analysis of the dopa decarboxylase gene cluster mutants in Drosophila melanogaster. J Hered. 1992;87:175–190.
- Noguchi H, Hayakawa Y. Dopamine is a key factor for the induction of egg diapause of the silkworm, Bombyx mori. Eur J Biochem. 2001;268:774–780.10.1046/j.1432-1327.2001.01933.x
- Yamashita O. Diapause hormone of the silkworm, Bombyx mori: structure, gene expression and function. J Insect Physiol. 1996;42:669–679.10.1016/0022-1910(96)00003-0
- Andersen SO. Insect cuticular sclerotization: a review. Insect Biochem Mol Biol. 2010;40:166–178.10.1016/j.ibmb.2009.10.007
- Lunan KD, Mitchell HK. The metabolism of tyrosine-O-phosphate in Drosophila. Arch Biochem Biophys. 1969;132:450–456.10.1016/0003-9861(69)90388-9
- Scholnick SB, Morgan BA, Hirsh J. The cloned dopa decarboxylase gene is developmentally regulated when reintegrated into the drosophila genome. Cell. 1983;34:37–45.10.1016/0092-8674(83)90134-4
- Johnson WA, McCormick CA, Bray SJ, et al. A neuron-specific enhancer of the Drosophila dopa decarboxylase gene. Genes Dev. 1989;3:676–686.10.1101/gad.3.5.676
- Morgan BA, Johnson WA, Hirsh J. Regulated splicing produces different forms of dopa decarboxylase in the central nervous system and hypoderm of Drosophila melanogaster. EMBO J. 1986;5:3335–3342.
- Hwang JS, Kang SW, Goo TW, et al. cDNA cloning and mRNA expression of L-3,4-dihydroxyphenylalanine decarboxylase gene homologue from the silkworm, Bombyx mori. Biotechnol Lett. 2003;25:997–1002.10.1023/A:1024035424317
