Abstract
Cell wall-associated β-xylosidase was isolated from Aspergillus niger E-1 and identified as XlsIV, corresponding to the extracellular enzyme XlnD reported previously. xlsIV was transcribed only in the early cultivation period. Cell wall-associated enzyme activity gradually decreased, but extracellular activity increased as the strain grew. These results indicate that XlsIV (XlnD) was secreted into culture after localizing at cell wall.
Filamentous fungi, Aspergillus is well known to produce β-xylosidase (EC 3.2.1.37).Citation1) Among Aspergillus species, Aspergillus niger is also able to degrade xylan completely and possesses five putative β-xylosidase genes, which are needed in the final step of xylan degradation.Citation2) Cloning and characterization of xlnD, which encodes the major extracellular β-xylosidase in A. niger, revealed that XlnD plays essential roles in the complete hydrolysis of xylan.Citation3,4) Although some researchers have reported the production of extracellular or intracellular β-xylosidases and their enzymatic properties,Citation5–7) the genes encoding these β-xylosidases are unknown. Thus, studies of the expression of A. niger β-xylosidase genes and the localization of translated enzymes remain still limited. In a previous study, we isolated A. niger E-1 as an excellent xylan degrader.Citation8) We describe here the localization of β-xylosidases produced by A. niger E-1 and their gene expression.
The first, we confirmed the presence of β-xylosidase genes in A. niger E-1 genome by polymerase chain reaction (PCR) using primer pairs designed on the basis of A. niger CBS513.88 β-xylosidase gene sequences.Citation2) For the preparation of total DNA, A. niger E-1 was cultivated in PGY medium (pH 6.0).Citation8) Total DNA was extracted by the method used in a previous study.Citation9) PCR was performed using MightyAmp Taq DNA polymerase (Takara Bio, Ohtu, Japan) or, if necessary, KOD DNA polymerase (Toyobo, Osaka, Japan), and primers (Table S1). The amplified fragments were cloned and sequenced as previously described.Citation9) DNA fragments amplified with KOD DNA polymerase were directly sequenced after purification. A. niger E-1 also harbored five putative β-xylosidase genes and these were designated as xlsI–xlsV in the ascending order of molecular masses calculated from the deduced amino acid sequences. The DDBJ/EMBL/GenBank accession numbers for the sequences described here are LC077866–LC077870. Their nucleotide sequences showed 95.4–99.9% identities with those of each corresponding gene in strain CBS513.88 genome (Table S2).
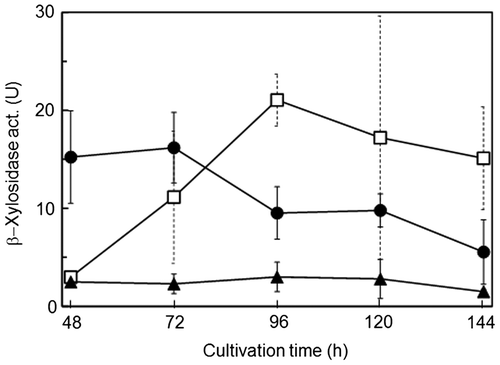
To elucidate the localization of β-xylosidases from A. niger E-1, the strain was inoculated with spores (5 × 106) in 50 mL of 0.5% beechwood xylan medium (pH 5.5) supplemented with 50 mM succinateCitation9) (XS medium) and cultivated at 37°C with shaking. The culture was filtered after being cultivated for 48, 72, 96, 120, and 144 h. The filtrates were centrifuged and kept as extracellular enzyme fractions. The harvested mycelia were used for the preparation of cell wall-associated and intracellular enzyme fractions. Cell wall-associated enzyme fractions were prepared according to the methods of Iwashita et al.Citation10) with slight modifications. The mycelia (0.1 g) washed with sterilized water were incubated in 1 mL of isotonic buffer (0.8 M NaCl–20 mM acetate buffer, pH 5.5) containing 5 mg of Yatalase™ (Takara Bio) for 2 h at 37°C. After centrifugation, the supernatants were used as cell wall-associated enzyme fractions. The precipitates were washed with 1.5 mL of isotonic buffer three times and resuspended in 1 mL of 20 mM Tris–HCl buffer (pH 7.5). The suspended cells were disrupted three times by a Mixer Mill MM2 (Retche, Haan, Germany) for 2 min with zirconia beads (6 mm in diameter) at 2 min intervals on ice. After centrifugation, the supernatants were used as intracellular enzyme fractions. β-Xylosidase activity was measured by the previous methods.Citation8) One unit (U) of β-xylosidase activity was defined as the amount of enzyme that releases 1 μmol of p-nitrophenol as a xylose equivalent per minute. As shown in Fig. , cell wall-associated β-xylosidase activity decreased with culture time, whereas extracellular enzyme activity tended to increase until 96 h and decreased gradually. There was little activity in the intracellular enzyme fractions.
Fig. 1. Localization of β-xylosidases produced by Aspergillus niger E-1.
Extracellular enzyme fraction (63 U, 144 mL) prepared from 96-h cultivated filtrate was dialyzed twice against 2 L of 20 mM acetate buffer (pH 5.5; buffer A) and applied to a DEAE-Sepharose Fast Flow (GE Healthcare, Buckinghamshire, UK) anion-exchange column (1.6 × 14.5 cm) equilibrated with buffer A at a flow rate of 50 mL/h. Proteins were eluted with a linear gradient of NaCl (0–0.5 M) at the same flow rate, and the eluate was separated into tubes (3.8 mL/tube) for β-xylosidase assay. All procedures were performed at 4°C. Protein concentrations were estimated by a DC™ Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA). One active peak (peak A; 61 U), eluted at the concentration of 0.19 M NaCl, was observed in the extracellular enzyme fraction (Fig. S1). Proteins contained in the fraction were separated by 7.5% (w/v) SDS-PAGECitation11) (Fig. ) and analyzed using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) according to the previous methods.Citation9) A protein band (EX-I) with the molecular mass of 121 kDa was identified as exo-1,4-β-xylosidase, XlnD with a protein score of 71. XlnD, corresponding to XlsIV in strain E-1, has been reported to be major extracellular β-xylosidase of A. niger in previous studies.Citation3,4)
Fig. 2. SDS-PAGE of β-xylosidase preparations.
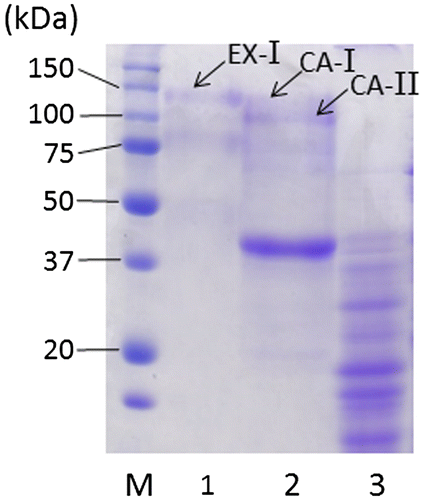
A cell wall-associated enzyme fraction (12 U, 4.7 mL) was prepared from 0.5 g of 48-h-grown mycelia and analyzed by the same methods described above after dialysis against 1 L of buffer A. Proteins were eluted with a linear gradient of NaCl (0–0.3 M) and one active peak (peak B; 8 U) was found in a fraction eluted at 0.18 M NaCl concentration (Fig. S2). SDS-PAGE analysis revealed that peak B contained two protein bans (CA-I and CA-II) with molecular masses (118 and 98 kDa, respectively) similar to that of EX-I (Fig. ). CA-I and CA-II were also identified as XlnD (XlsIV in strain E-1) using MALDI-TOF-MS. To our knowledge, this is the first report that cell wall-associated β-xylosidase is XlnD and that both extracellular and cell wall-associated β-xylosidases are encoded by the same gene. MALDI-TOF-MS analysis of other bands in the cell wall-associated enzyme fraction also suggested that a predominant band (39 kDa) was not, at least, β-xylosidases and other plant fiber-degrading enzymes from A. niger (data not shown) and that proteins with β-xylosidases activity were derived from only CA-I and CA-II. The molecular masses of three kinds of XlsIV, obtained from extracellular and cell wall-associated fractions, were 98–121 kDa, whereas that of native XlnD has been reported to be 110 kDa and decreased to approximately 85 kDa after N-glycanase treatment.Citation3) From these studies, XlsIV also seem to be modified by N-glycosylation. Difference in molecular masses depending on localization has been also observed about β-glucosidases from Aspergillus kawachii.Citation10) A. kawachii produces two extracellular (EX-1 and EX-2) and one cell wall-bound (CB-1) β-glucosidases with molecular masses of 145, 130, and 120 kDa, respectively. This difference is attributed to the degree of N-link glycosylation for each enzyme. Interestingly, molecular masses of extracellular enzymes are bigger than that of the cell wall bound enzyme as well as our case. Thus, the degree of N-link glycosylation of Aspergillus plant fiber-degrading enzymes, such as β-glucosidases and β-xylosidases, may be regulated by unknown mechanisms depending on their localization.
Reverse transcription (RT)-PCR analysis was performed to identify the expressed β-xylosidase genes. Mycelia were cultivated in XS medium and harvested after 48-, 72-, 96-, and 120-h cultivation as described above. Total RNA was extracted from the mycelia (0.65 g in total) using zirconia beads (6 mm), ISOGEN (NIPPON GENE, Tokyo), and a Mixer Mill MM2. mRNA was obtained from total RNA using a PolyATract® mRNA Isolation System III (Promega, Madison, WI). RT reaction mixtures were prepared from the mRNA mixtures, upstream, and downstream primers specific for each β-xylosidase gene (Table S3), and an AccessQuick™ RT-PCR System (Promega) according to the supplier’s protocol and incubated at 45°C for 45 min. PCR was performed as follows: an initial denaturation step at 95°C for 2 min, followed by 41 cycles of denaturing step at 95°C for 45 s, annealing at 53°C for 45 s, and extension at 72°C for 3 min, and the final extension of for 5 min at 72°C. Amplified cDNA was analyzed using 2% agarose gel electrophoresis and directly sequenced using primers shown in Table S3 to confirm the sequences. The expression profile is shown in Fig. . The expression of xlsIV (panel A and C in Fig. ) was detected in the initial (48 h) to middle cultivation (72 h) period and xlsII transcript (panel B and C) was found at a later cultivation stage, 96–120 h. xlsI, xlsIII, and xlsV were not expressed at any time in this experiment.
Fig. 3. Expression profile of β-xylosidase genes from A. niger E-1.
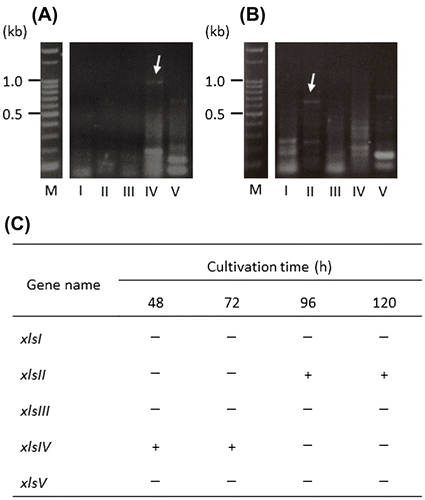
We prepared individual enzyme fractions from one culture to clarify the localization of β-xylosidases from A. niger E-1. As shown in Fig. , cell wall-associated β-xylosidase activity decreased, whereas extracellular enzyme activity increased with growth of the strain. A similar observation was made in a study of extracellular and not cell-associated but intracellular β-xylosidase production by A. fumigatus.Citation12) However, the cell wall-associated β-xylosidase produced by A. oryzae, increased after 72-h cultivation in 2% xylan liquid medium.Citation13) These results, including ours, suggest that the localization of β-xylosidase differs among fungal species, and varies with other factors such as growth phase and culture conditions. However, because it was unclear which β-xylosidase genes, whether single or multiple genes, encode β-xylosidases detectable at different locations, we attempted to identify both extracellular and cell wall-associated β-xylosidases from A. niger E-1 and found that both enzymes were XlsIV, corresponding to the exo-1,4-β-xylosidase XlnD. In addition, the expression of xlsIV was detected in 48- and 72-h-grown mycelia, and not in the 96-h culture (Fig. ). In view of the localization of XlsIV, these results suggest that xlsIV is transcribed during the initial to middle period in culture and that most of the xlsIV gene product XlsIV is first localized at the cell wall and later secreted in medium. As a case similar to β-xylosidases in this study, β-glucosidases from A. kawachii, EX-1 and EX-2, and CB-1, encoded by the single β-glucosidase gene bglA, are localized in cultural supernatant and cell wall, respectively.Citation10,14) Interestingly, their localization is controlled by extracellular soluble polysaccharide (ESP); the β-glucosidase is easily absorbed to the cell wall of A. kawachii, but secreted into medium by the inhibition of absorption to cell wall in the presence of ESP with absorption ability to the β-glucosidases. The component of ESP is similar to that of the alkaline-soluble fraction from cell wall of other Aspergillus species.Citation15) Time-dependent increase of extracellular XlsIV, which is thought to be released from cell wall, may be also caused by possible ESP coming from materials of cell wall surface.
We newly found xlsII transcript in 96- and 120-h-grown mycelia (Fig. ). The expression of xlsII is an interesting finding, given that the physiological properties and localization of XlsII are still unknown. Further study will lead us to understanding of the physiological roles of each β-xylosidase.
Supplemental material
Supplemental material for this article can be accessed at http://dx.doi.org/10.1080/09168451.2016.1268040
Author contribution
Y.T. and S.M. designed the research plan. K.O. performed additional experiments which were pointed out by reviewers. K.I. and Y.T. cooperatively performed other experiments. Y.T. wrote the manuscript and S.M. checked the manuscript.
TBBB_1268040_Supplementary_Materials.docx
Download MS Word (146.9 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Wakiyama M, Yoshihara K, Hayashi S, et al. Purification and properties of an extracellular β-xylosidase from Aspergillus japonicus and sequence analysis of the encoding gene. J Biosci Bioeng. 2008;106:398–404.10.1263/jbb.106.398
- Pel HJ, de Winde JH, Archer DB, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231.10.1038/nbt1282
- van Peij NNME, Brinkmann J, Vrsanska M, et al. β-xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173.10.1111/ejb.1997.245.issue-1
- Benassi VM, da Silva TM, Pessela BC, et al. Immobilization and biochemical properties of a β-xylosidase activated by glucose/xylose from Aspergillus niger USP-67 with transxylosylation activity. J Mol Catal B: Enzym. 2013;89:93–101.10.1016/j.molcatb.2012.12.010
- Uchida H, Kusakabe I, Kawabata Y, et al. Production of xylose from xylan with intracellular enzyme system of Aspergillus niger 5–16. J Ferment Bioeng. 1992;74:153–158.10.1016/0922-338X(92)90075-6
- Pedersen M, Lauritzen HK, Frisvad JC, et al. Identification of thermostable β-xylosidase activities produced by Aspergillus brasiliensis and Aspergillus niger. Biotechnol Lett. 2007;29:743–748.10.1007/s10529-007-9314-9
- Díaz-Malváez FI, García-Almendárez BE, Hernández-Arana A, et al. Isolation and properties of β-xylosidase from Aspergillus niger GS1 using corn pericarp upon solid state fermentation. Process Biochem. 2013;48:1018–1024.10.1016/j.procbio.2013.05.003
- Takahashi Y, Nomura M, Komiya T, et al. Screening of lignocelluloce-degrading fungi from gray gentle lemur (Hapalemur griseus) feces. Bull Sch Agric Meiji Univ. 2012;61:77–86.
- Takahashi Y, Kawabata H, Murakami S. Analysis of functional xylanases in xylan degradation by Aspergillus niger E-1 and characterization of the GH family 10 xylanase XynVII. SpringerPlus. 2013;2:447.10.1186/2193-1801-2-447
- Iwashita K, Todoroki K, Kimura H, et al. Purification and characterization of extracellular and cell wall bound β-glucosidases from Aspergillus kawachii. Biosci Biotechnol Biochem. 1998;62:1938–1946.10.1271/bbb.62.1938
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685.10.1038/227680a0
- Lenartovicz V, de Souza CGM, Moreira FG, et al. Temperature and carbon source affect the production and secretion of a thermostable β-xylosidase by Aspergillus fumigatus. Process Biochem. 2003;38:1775–1780.10.1016/S0032-9592(02)00261-3
- Hashimoto T, Morishita M, Iwashita K, et al. Production and some properties of salt-tolerant β-xylosidases from a Shoyu Koji Mold, Aspergillus oryzae in solid and liquid cultures. J Biosci Bioeng. 1999;88:479–483.10.1016/S1389-1723(00)87662-8
- Iwashita K, Nagahara T, Kimura H, et al. The bglA gene of Aspergillus kawachii encodes both extracellular and cell wall-bound β-glucosidases. Appl Environ Microbiol. 1999;65:5546–5553.
- Iwashita K, Shimoi H, Ito K. Extracellular soluble polysaccharide (ESP) from Aspergillus kawachii improves the stability of extracellular β-gluocosidases (EX-1 and EX-2) and is involved in their localization. J Biosci Bioeng. 2001;91:134–140.10.1016/S1389-1723(01)80055-4
