Abstract
The function of microRNA-34a (miR-34a) in transdifferentiation of glioma stem cells (GSCs) into vascular endothelial cells (VECs) was explored by focusing on Notch ligand Delta-like 1 (Dll1). MiR-34a mimics was transfected into CD133 + glioma cell U251. The angiogenesis feature of miR-34a transfected U251 cells was investigated and the expressions of CD31, CD34, Vwf, Notch 1, and Dll1 were quantified. Length of branching vessel-like structures in the miR-34a transfected U251 cells was significantly higher than control cells. The VEC feature of miR-34a overexpressed U251 cells was further confirmed by the expressions of CD31, CD34, and vWF. Transfection of miR-34a decreased the expression of Notch 1 and Dll1. Furthermore, the miR-34a overexpression-enhanced tube formation of GSCs was suppressed when the decreased expression of Dll1 was restored. The current study highlighted the potential of miR-34a as an inducer in GSCs’ transdifferentiation into VECs by targeting Dll1.
Up-regulation of miR-34a induced transdifferentiation of CD133+ U251 cells into VECs
The theory that tumor is sustained by a subpopulation of cells defines the concept of cancer stem cells (CSCs).Citation1) CSCs, characterized by self-renewal and generation of a heterogeneous mass of tumor cells, have been demonstrated in glioma, namely, glioma stem cells (GSCs). Glioma is the most prevalent and devastating type of primary brain tumor.Citation2) In clinic, this malignancy is conventionally managed by maximal surgical resection followed by radio and chemotherapy.Citation3,4) However, due to the high recurrence of the tumor, glioma patients treated with rigorous surgery and chemotherapy merely have a median survival time of 15–19 months.Citation5) Additionally, emerging evidence suggests that GSCs are responsible for the obstacles in the treatment and prognosis of glioma as they are highly resistant to radio and chemotherapy.Citation6) Thus, a comprehensive understanding of GSC-involved glioma progression would help the development of novel anti-glioma therapies.
GSCs are capable of transdifferentiating into vascular endothelial cells (VECs) and participate in the neovascularization process of glioma.Citation7–9) Endothelial cells play a central role in the progression of malignant glioma as they create a niche for GSCs to reside in and self-renew.Citation10) The so-called “stem cell niche” in glioma consists of GSCs, neighboring supportive cells, extracellular matrix, and other factors necessary for stem cell self-renewal.Citation11) A recent study by Ricci-Vitiani et al. demonstrated that selective inhibition of GSC-derived VECs suppressed the growth of tumor xenografts in mice, presenting the functional association of GSC-derived endothelial vessels with the tumorigenesis of glioma.Citation12) Moreover, endothelial cells are likely to promote self-renewal of GSCs by providing Notch ligands.Citation10) The Notch pathway is an evolutionarily conserved intercellular signaling pathway that plays important roles in the oncogenesis and development of glioma.Citation13) Expression of the constitutively active intracellular domains of Notch 1 or Notch 2 increases the resistance of GSCs to radiotherapy,Citation14) and down-regulation of Notch 1 inhibits the proliferation of glioma cells.Citation15) In addition, the Notch pathway also regulates tumor cell’s transdifferentiation into endothelial cells in multiple mannersCitation16–18): activation of Notch-1 by Delta like 4 (Dll4) reduces tumor vascularityCitation19) while Jagged1 has been described as a Notch ligand expressed in tumor cells that can have a positive influence on tumor angiogenesis.Citation18,20) Thus, specifically targeting the Notch signaling pathway may provide novel targets for the development of anti-glioma therapies.
The miR-34 family members, including miR-34a, miR-34b, miR-34c, are identified as the miR components of the p53 network because their expressions are closely correlated with p53.Citation21) De Antonellis et al. have demonstrated that miR-34a modulates the activity of Notch signaling by specifically targeting the Notch ligand Dll1 and thus influences the neural differentiation of medulloblastoma cells.Citation15,22) Additionally, Luan and his colleagues confirm the tumor suppressor activity of miR-34a in U251 glioma cell line.Citation21) Given that Dll1’s function in the vasculature was less studied, it will be interesting to determine role of miR-34a as well as its interaction with Dll1 in glioma neovascularization. In the current study, expression of exogenous miR-34a in CD133 + U251 cells (considered as GSCs) was induced in order to investigate the function of miR-34a in the transdifferentiation of GSCs into VECs via Notch signaling. The findings outlined in the current study revealed that miR-34a may induce transdifferentiation of GSCs into VECs by inhibiting the Notch signaling pathway.
Materials and methods
Reagents and cells
Specific mimics of miR-34a (5′-UGGCAGUGUCUUAGCUGGUUGU-3′) and scramble negative control (NC) oligonucleotides (5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from GenePharma (Shanghai, China). Antibody against CD133 (Catalog. No. 130-098-826) was purchased from Miltenyl Biotec (Miltenyi Biotec, Bergisch Gladbach, FRG). Antibodies against Nestin (Catalog. No. sc-20978) and β-actin (Catalog. No. sc-47778) were purchased from Biotechnology, Inc (Santa Cruz, CA). Antibodies against CD31 (Catalog. No. BA1346), CD34 (Catalog. No. BA0532), and vWF (Catalog. No. PB0086) were purchased from BOSTER (BOSTER, China). Antibody against Notch 1 (Catalog. No. D161041) was purchased from Sango Biotech (Sangon Biotech, China). Antibody against Dll1 (Catalog. No. bs-7345R) was purchased from Beijing Biosynthesis Biotechnology Co., LTD (Biosynthesis Biotechnology, China). Human U251 glioma cell line (Catalog. No. TCHu 58) was obtained from Shanghai Cell Bank of Chinese Academy of Sciences, and cultured in DMEM medium supplemented with 10% FBS. The cells were adjusted to 1 × 108/mL and labeled using the Miltenyi Biotec CD133 MicroBead Kit (Catalog. No. 130-097-049, Miltenyi Biotec, Bergisch Gladbach, Germany). The sorted CD133 + and nestin + U251 cells that displayed neurosphere-like morphology, nonadherent growth, and asymmetrical cell divisions were further identified by immunofluorescent assay.Citation23) For the subsequent assays, CD133 + U251 cells were cultured in the medium containing DMEM/F12, 100 U/mL benzylpenicillin, 100 μg/mL streptomycin, 20 μL/mL B27, 20 ng/mL EGF, and 20 ng/mL bFGF in an atmosphere of 5% CO2 and saturated humidity at 37 °C. The cells from passages three to five were used for the subsequent assays.
Construction of Dll1 expression vector and cell transfection
To assess the function of miR-34a in the transdifferentiation of GSCs into VECs, CD133 + U251 cells were divided into three groups with different treatments: (A) Control group, untreated CD133 + U251 cells. (B) NC group, CD133 + U251 cells transfected with the scramble NC oligos. (C) miR-34a group, CD133 + U251 cells transfected with miR-34a mimics. The role of Dll1 in the function of miR-34a was detected by ligating the CDS of Dll1 gene into expression plasmid and the CD133 + U251 cells were grouped as following: (A) miR-34a group, CD133 + U251 cells transfected with miR-34a mimics. (B) Vector group, CD133 + U251 cells co-transfected with miR-34a mimics and NC plasmid. (C) Dll1 group, CD133 + U251 cells co-transfected with miR-34a mimics and Dll1 expression plasmid. Transfection was performed using Lipofectamine2000 (Catalog. No. 11668-019, Invitrogen) according to the manufacturer’s instructions. At 48 h post-transfection, the cells were collected for the subsequent assays and each following assay was represented by at least three replicates.
Immunofluorescent detection
Immunofluorescent detection was conducted following previous studyCitation24): briefly, paraformaldehyde fixed cells were permeabilized with 0.1% Triton X-100 for 30 min. After being blocked in 10% goat serum for 15 min, primary rabbit polyclonal antibodies against different proteins of interest (1:200) (CD133, Nesting, CD31, CD34, vWF, Notch or Dll1) were then added and the cells were incubated overnight at 4 °C and then incubated with different Cy3-labeled secondary antibodies (1:200) for 1 h in dark. Thereafter, the cells were stained with 4,6-diamino-2-phenyl indole (DAPI) for 5 min at room temperature. The results were imaged with the fluorescent microscope (BX53, OLYMPUS) at 400× magnification.
Tube formation assay
Three-dimensional culture was performed according to study of Zhao et al.Citation9) with some modifications: a 24-well dish was pre-coated with matrigel (Catalog. No. 40111ES08, BD bioscence, USA) at 50 μL/well and placed in a 37 °C incubator with a humidified atmosphere of 5% CO2 and 95% air for gelation for 2 h. CD133 + U251 cells (1 × 104 cells/well), suspended in 100 μL endothelial differentiation medium, were plated into each well and allow to growth for 8 days. Cell morphology was observed with a Nikon inverted microscope (Eclipse TE2000-U) at 100× magnification.
RNA extraction and reverse transcription PCR (RT-PCR)
Total RNAs were extracted using RNA extraction kit according to the manufacturers’ instruction (Catalog. No. RP1201, BioTeke, China). cDNA templates were obtained via reverse transcription of the RNA using Super M-MLV reverse transcriptase (Catalog. No. PR6502, BioTeke, China). The final reaction mixture of volume 20 μL contained 10 μL of 2 × Taq PCR Master-mix (Catalog. No. PR1702, BioTeke, China), 1 μL of each primers [CD31 forward: 5′-CACCGAGTTGTATCACGAAA-3′, reverse: 5′-TAAGGCACCAGGAGTAGGAA-3′. CD34, forward: 5′-GTCTTCCACTCGGTGCGTCTC-3′, reverse: 5′-GTTCCCTGGGTAGGTAACTCT-3′. vWF, forward: 5′-TCCACCGAAGCACCATCTA-3′, reverse: 5′-AGCCAGTCACCACCTCACA-3′. β-actin (internal reference) forward: 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′, reverse: 5′- TGTCACCTTCACCGTTCCAGTTT-3′], 1 μL of the cDNA template, and 7 μL of Rnase free H2O. Thermal cycling parameters for the amplification were as follows: a denaturation step at 95 °C for 5 min, followed by 36 cycles at 95 °C for 20 s, 52 °C for 20 s, and 72 °C for 30 s, and the reaction was stopped by a step at 25 °C for 5 min. The images of gels of CD31, CD34, vWF were recorded with a Gel imaging analyzer (WD-9413B, Liuyi, Beijing, China).
Reverse transcription real-time PCR (RT2-PCR)
The final reaction mixture of volume 20 μL contained 10 μL of SYBR GREEN mastermix (Catalog. No. SY1020, Solarbio, China), 1 μL of each primers [Notch 1, forward: 5′-CTGCCCGTTCTTGAAATGTAGG-3′, reverse: 5′-TGTTGCTGGAGCATCTTCTTCG-3′. Dll1, forward: 5′-GGACTCGGGCTGTTCAACTT-3′, reverse: 5′-CTTGCCATCTCACTTCCATT-3′. β-actin (internal reference) forward: 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′, reverse: 5′- TGTCACCTTCACCGTTCCAGTTT-3′]. Thermal cycling parameters for the amplification were as follows: a denaturation step at 95 °C for 5 min, followed by 36 cycles at 95 °C for 20 s, 62 °C for 20 s, and 72 °C for 30 s, and the reaction was stopped by a step at 4 °C for 5 min. The relative expression levels of Notch1 and Dll1 were detected by real time PCR system (ExicyclerTM 96, BIONEER, South Korea) according to the expression of 2−△△ct.
Western blotting assay
Total proteins were extracted using the total protein extraction kit according to the manufacturer’s instructions (Catl. No. WLA019, Wanleibio, China). The concentrations of the protein samples were determined according to the BCA method using a BCA assay kit purchased from Wanleibio (Catl. No. WLA004, Wanleibio, China). Western blotting assay was performed following standard procedures: briefly, the membranes were incubated with one of the primary antibodies against CD31 (1:400), CD34 (1:300), vWF (1:400), Notch 1 (1:500), Dll1 (1:500), and β-actin (internal reference) (1:1000) overnight at 4 °C and the then incubated with peroxidase-conjugated secondary antibodies (1:5000). The blotting images were analyzed with the gel imaging system (WD-9413B, Liuyi, Beijing, China) and the relative expression levels of different proteins were calculated with bio-rad quantity one software v. 4.6.3 (Bio-Rad).
Statistical analysis
All the data were expressed as mean ± SD (n = 3). One-way ANOVA and post hoc comparisons using LSD method were conducted with different assays. All the statistical analyses were performed using R language version 3.2.1, and the images were edited using ggplot2 package 3.1.1 in R language (R foundation for statistical computing).
Results
Up-regulation of miR-34a induced tube formation in CD133 + U251 cells
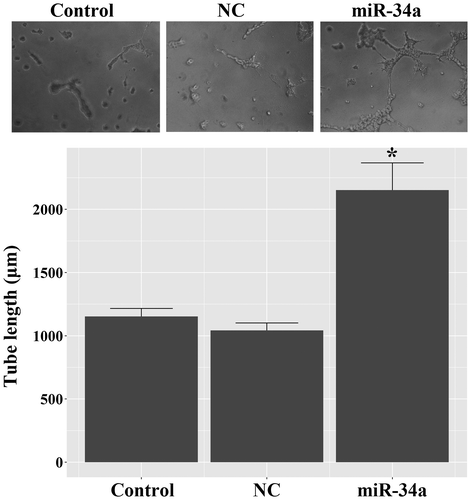
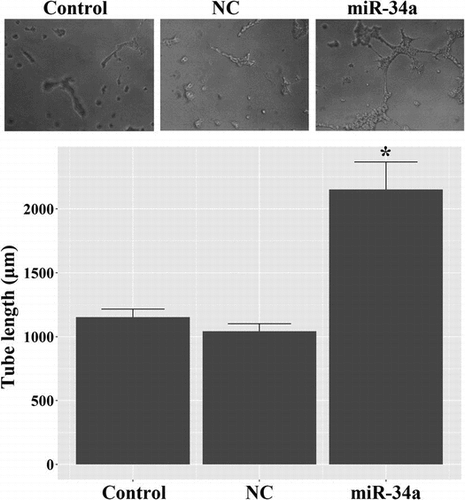
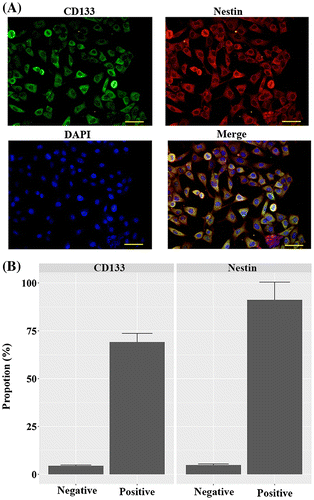
CD133 + U251 cells were isolated from U251 glioma cells by employing immunomagnetic beads, and the sorted cells were characterized by positive staining for CD133 and Nestin (Fig. ). As shown in the Fig. , the high proportions of CD133 + and Nestin + cells indicated that the sorted population was GSC-enriched cells. Next, tube formation assay was conducted to assess the potential of miR-34a in regulating endothelial fate determination of GSCs. As shown in Fig. , miR-34a mimics-induced evident tube formation in CD133 + U251 cells: miR-34a-transfected cells showed branching vessel-like structures after 8 days of culture in matrigel. The overall tube length in miR-34a-transfected CD133 + U251 cells was higher than those in the Control and NC cells, and the differences were statistically significant (p < 0.05). This result demonstrated the potential of miR-34a in inducing GSCs’ transdifferentiation into VECs.
Fig. 1. Identification of the stem cell features of CD133 + U251 cells by immunofluorescent assay. CD133 + cells were isolated from U251 cells using anti-CD133 immunomagnetic beads, and the sorted cells were positively stained for CD133 and Nestin, representing stem cell-like characteristics. (A) representative images of immunofluorescent staining of CD133 and Nestin in the sorted cells. (B) quantitative analysis of the proportions of CD133 + and Nestin + cells in the sorted population. Scale bar, 50 μm.
Up-regulation of miR-34a induced transdifferentiation of CD133 + U251 cells into VECs
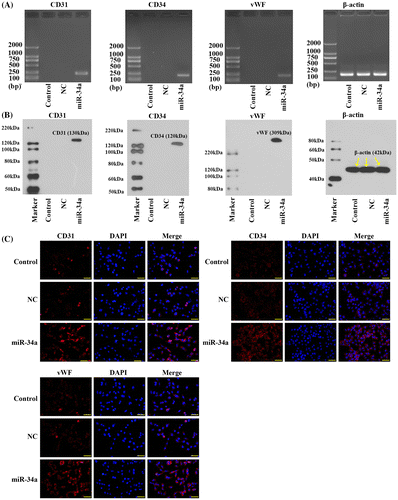
The endothelial nature of miR-34a-transfected CD133 + U251 cells was further assessed by detecting the expressions of three endothelial markers, CD31, CD34, and vWF, using RT-PCR, western blotting assay, and immunofluorescent detection. None of the endothelial markers was detected in the Control and NC groups at either mRNA or protein level (Fig. (A), (B)), indicating the absence of endothelial markers in the native CD133 + U251 cells. Similar results were observed in the immuonfluorescent assay (Fig. (C)). After transfection with miR-34a mimics, the cells showed evident expressions of the endothelial markers (Fig. ). These results confirmed the endothelial nature of the GSCs transfected with miR-34a.
Fig. 3. Transfection with miR-34a mimics induced the expression of endothelial markers in CD133 + U251 cells. (A) Representative images of RT-PCR validation of the expression of endothelial markers: the expressions of CD31, CD34, and vWF were observed only in the CD133 + U251 cells transfected with miR-34a. (B) Representative images of western blotting validation of expressions of endothelial markers: the expressions of CD31, CD34, and vWF were observed only in the CD133 + U251 cells transfected with miR-34a. (C) Representative images of immunofluorescent staining of the three endothelial markers: more immunofluorescent positive cells were observed in CD133 + U251 cells transfected with miR-34a. Scale bar, 50 μm.
miR-34a down-regulated the Notch pathway components during transdifferentiation of GSCs into VECs
The Notch pathway is a central signaling cascade in the regulation of tumor angiogenesis. In the current study, the effect of miR-34a on the expressions of Notch 1 and its ligand Dll1 was investigated. As shown in Fig. (A), the level of Notch 1 mRNA was down-regulated by miR-34a mimics, and the differences between the miR-34a group and the other two groups were statistically significant (p < 0.05). Concomitant with the reduction of Notch 1 mRNA, the level of Dll1 mRNA was also significantly decreased in miR-34a-transfected cells (p < 0.05). Consistent with the RT2-PCR results, western blotting showed reduced expression levels of Notch 1 and Dll1 proteins in miR-34a-transfected cells (Fig. (B); p < 0.05). Furthermore, the inhibitory effect of miR-34a on the expressions of Notch1 and Dll1 was validated by immunofluorescent assay: Notch 1 and Dll1 proteins in CD133 + U251 were markedly decreased after transfection with miR-34a mimics (Fig. ).
Fig. 4. Transfection with miR-34a mimics down-regulated Notch pathway components in CD133 + U251 cells. (A) Quantitative RT2-PCR analysis of the expression levels of Notch 1 and Dll1. (B) Representative images and quantitative analysis of western blotting assay of the expressions of Notch 1 and Dll1. “*” represents a significant difference from the Control and NC groups, p < 0.05.
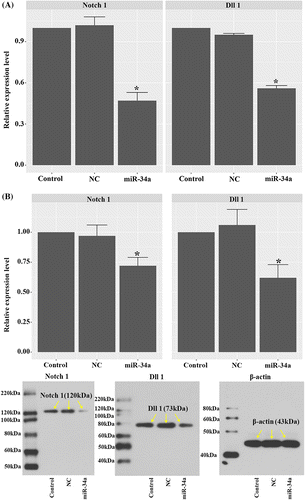
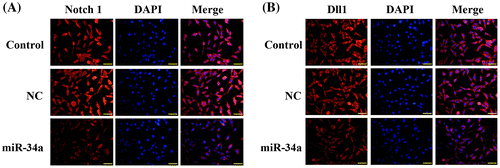
Fig. 5. Transfection with miR-34a mimics inhibited the expressions of Notch 1 and Dll1 as validated by immunofluorescent assay. (A) Representative images of immunofluorescent staining of Notch 1 in CD133 + U251 cells transfected with miR-34a mimics or NC oligos. (B) Representative images of immunofluorescent staining of Dll1 in different groups. Scale bar, 50 μm.
Overexpression of Dll1 counteracted the effect of miR-34a mimics to induce vasculature transdifferentiation of CD133 + U251 cells by activating Notch 1
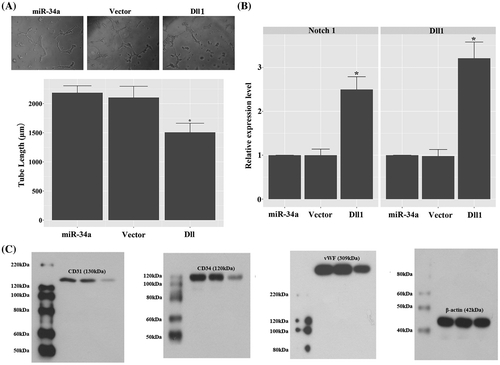
To explicitly demonstrate the role of Dll1 in the function of miR-34a in GSCs’ vasculature transdifferentiation, the expression of Dll1 was induced in miR-34a mimics transfected CD133 + U251 cells. Our data showed that increased expression of Dll1 inhibited the tube formation induced by miR-34a mimics: the tube length in Dll1 group was shorter than that in miR-34a group, and the difference was statistically significant (p < 0.05; Fig. (A)). The increased expression of Dll1 also lead to up-regulation of Notch 1 (Fig. (B)), which would inhibit the neovascularization of tumors.Citation19) Furthermore, the neovascularization inhibition effect of Dll1 overexpression was confirmed by decreased expression of CD31, CD34, and vWF which are classical markers of VECs (Fig. (C)).
Fig. 6. Overexpression of Dll1 confounded the effect of miR-34a mimics to induce vasculature transdifferentiation of CD133 + U251 cells by inducing the expression of Notch 1. (A) Overexpression of Dll1 significantly inhibited tube formation in miR-34a mimics transfected CD133 + U251 cells. 100×. (B) Quantitative RT2-PCR analysis of the expression levels of Notch 1 and Dll1. (C) Representative images of western blotting validation of expressions of endothelial markers: the expressions of CD31, CD34, and vWF were inhibited in the CD133 + U251 cells transfected with miR-34a mimics and Dll1 expression vector. “*”, significantly different from miR-34a and vector groups, p < 0.05.
Discussion
MiR-34a is recognized as a potential tumor suppressor,Citation25) and it has become a hot subject of cancer-related studies in the past decade.Citation21,26,27) The level of miR-34a is shown to be positively correlated with the expression of p53 in medulloblastoma and glioblastoma multiforme.Citation28,29) Loss of miR-34a presents a mechanism through which p53 pathway is inhibited and chemosensitivity of malignant glioma is modulated.Citation21) On the basis of these studies, increasing efforts have been made to evaluate the potential of miR-34a as a therapeutic target for glioma.
Currently, promising progress has been achieved by numerous studies focusing on the therapeutic value of miR-34a in glioma. For example, miR-34a has been demonstrated to inhibit the growth of glioblastoma by targeting c-Met and Notch.Citation30) MiR-34a also acts as a tumor suppressor in p53-mutant glioma U251 cells through the regulation of SIRT1.Citation21) In addition, as a multiple functional miR, miR-34a not only influences cell cycle and apoptosis, but also plays an important role in neovascularization.Citation31) Considering that the critical role of neovascularization in tumor development is well recognized, the present study aimed to explore the function of miR-34a in the transdifferentiation of GSCs into VECs. Our work demonstrated that up-regulation of miR-34a in CD133 + U251 cells, the GSCs, induced differentiation of the cells into VECs by targeting the Notch signaling pathway.
In the current study, CD133 + U251 cells were sorted by immunomagnetic bead-aided separation. Although glioma cells display a similar morphological phenotype, the biological features such as growth and differentiation capabilities vary among individual cells. Additionally, the CD133 + subpopulation in glioma cells suggests the presence of stem cell-like tumor cells,Citation32) and the neural stem-like feature of CD133 + U251 cells were further supported by the positive staining for Nestin, a hallmark of GSCs.Citation33)
To investigate the role of miR-34a in the transdifferentiation of GSCs toward VECs, miR-34a mimics was transfected into CD133 + U251 cells, and the phenotypic changes in the cells were examined by an in vitro vascularization assay. The results showed that miR-34a-induced tube formation in CD133 + U251 cells, displaying an endothelial phenotype. In addition, miR-34a-transfected CD133 + U251 cells showed evidently increased expressions of CD31, CD34, and vWF, all of which are typical markers of VECs.Citation9,34) These results demonstrated the commitment of GSCs to the VEC lineage by induced expression of miR-34a. Moreover, miR-34a mimics markedly decreased the expressions of Notch 1 and its ligand Dll1 in CD133 + U251 cells as detected by western blotting and immunofluorescent staining. As previously reported, miR-34a-mediated down-regulation of Notch 1 contributed to the inhibition of cell proliferation and tumor formation.Citation35) In addition, direct targeting of Dll1 by miR-34a supported neural differentiation of CD15+/CD133 + tumor-propagating cells in medulloblastoma.Citation22) A similar mechanism could be established in glioma based on the results of our study: by directly repressing the expression of Dll1, miR-34a induces the differentiation of GSCs toward VECs, while miR-34a-induced down-regulation of Notch 1 may also lead to the suppression of glioma development. Moreover, induced expression of Dll1 could counteract the effect of miR-34a mimics on CD133 + U251 cells, inhibiting tube formation and expression of CD31, CD34, and vWF, and activating Notch signaling induced by miR-34a overexpression. However, once committed to VECs, GSC-derived cells would subsequently initiate neovascularization in glioma, which is always associated with poor prognosis in glioma patients.Citation12,36) Thus, the VEC-inducing activity of miR-34a seems conflict with its proposed tumor repressor function. Contrary to our observations in glioma-derived VECs, miR-34a was previously shown to induce endothelial progenitor cell senescence and inhibit angiogenesis.Citation31) Moreover, the inhibitory effect of miR-34a on Notch 1 in our study also contradicts to the results from de Antonellis et al. that miR-34a-mediated inhibition of Dll1 leads to the activation of Notch 1 in medulloblastoma.Citation22) Therefore, although our work demonstrated that miR-34a promoted transdifferentiation of GSCs into VECs, the complicated and seemingly cell type-specific mechanism underlying the function of miR-34a in tumors demands further investigation.
In summary, the current study demonstrated a key role of miR-34a in the transdifferentiation of GSCs into VECs. During this process, miR-34a might exert its function through the inhibition of Notch signaling pathway. The VEC-inducing effect of miR-34a on GSCs might confound the function of miR-34a as a glioma suppressor. Therefore, the potential of miR-34a in treating glioma should be re-considered and comprehensively addressed in future studies.
Author contributions
Zaishun Jin and Tao Zhan designed the experiment, collected the data, and wrote the draft of the work. Jing Tao, Biao Xu, Huizhe Zheng, and Yongxia Cheng collected data. Bin Yan, Hongwei Wang, and Guoqiang Lu performed the analysis. Ying Lin and Sufen Guo revised the draft. Sufen Guo is responsible for the project.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the Natural Science Foundation of Heilongjiang Province [grant number H201492].
References
- Sampetrean O, Saya H. Characteristics of glioma stem cells. Brain Tumor Pathol. 2013;30(4):209–214.10.1007/s10014-013-0141-5
- Weller M. Novel diagnostic and therapeutic approaches to malignant glioma. Swiss Med Wkly. 2011;141:w13210.
- DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123.10.1056/NEJM200101113440207
- Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide vs. radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466.10.1016/S1470-2045(09)70025-7
- Grossman SA, Batara JF. Current management of glioblastoma multiforme. Semin Oncol. 2004;31(5):635–644.10.1053/j.seminoncol.2004.07.005
- Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760.10.1038/nature05236
- Hardee ME, Zagzag D. Mechanisms of glioma-associated neovascularization. Am J Pathol. 2012;181(4):1126–1141.10.1016/j.ajpath.2012.06.030
- Jhaveri N, Chen TC, Hofman FM. Tumor vasculature and glioma stem cells: contributions to glioma progression. Cancer Lett. 2014;380(2):545–551.
- Zhao Y, Dong J, Huang Q, et al. Endothelial cell transdifferentiation of human glioma stem progenitor cells in vitro. Brain Res Bull. 2010;82(5–6):308–312.10.1016/j.brainresbull.2010.06.006
- Zhu TS, Costello MA, Talsma CE, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71(18):6061–6072.10.1158/0008-5472.CAN-10-4269
- Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116(6):769–778.10.1016/S0092-8674(04)00255-7
- Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–828.10.1038/nature09557
- Lino MM, Merlo A, Boulay JL. Notch signaling in glioblastoma: a developmental drug target? BMC Med. 2010;8:72–80.10.1186/1741-7015-8-72
- Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28(1):17–28.
- Li WB, Ma MW, Dong LJ, et al. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol Ther. 2011;12(6):477–483.10.4161/cbt.12.6.16300
- Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer Cell. 2005;8(1):1.10.1016/j.ccr.2005.06.013
- Thurston G, Kitajewski J. VEGF and delta-notch: interacting signalling pathways in tumour angiogenesis. Br J Cancer. 2008;99(8):1204.10.1038/sj.bjc.6604484
- Rui B, Roca C, Sörensen I, et al. The notch ligands Dll4 and jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124.
- Segarra M, Williams CK, Sierra ML, et al. Dll4 activation of notch signaling reduces tumor vascularity and inhibits tumor growth. Blood. 2008;112(5):1904.10.1182/blood-2007-11-126045
- Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27(38):5132–5137.10.1038/onc.2008.227
- Luan S, Sun L, Huang F. MicroRNA-34a: a novel tumor suppressor in p53-mutant glioma cell line U251. Arch Med Res. 2010;41(2):67–74.10.1016/j.arcmed.2010.02.007
- De Antonellis P, Medaglia C, Cusanelli E, et al. MiR-34a targeting of notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE. 2011;6(9):e24584.10.1371/journal.pone.0024584
- Grosse-Gehling P, Fargeas CA, Dittfeld C, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol. 2013;229(3):355–378.10.1002/path.4086
- Yu H, Shi L, Qi G, et al. Gypenoside protects cardiomyocytes against ischemia-reperfusion injury via the inhibition of mitogen-activated protein kinase mediated nuclear factor kappa B pathway in vitro and in vivo. Front Pharmacol. 2016;7:148.
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022.10.1038/sj.onc.1210293
- Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215.10.1038/nm.2284
- Nalls D, Tang SN, Rodova M, et al. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6(8):e24099.10.1371/journal.pone.0024099
- Wang BC, Ma J. Role of MicroRNAs in malignant glioma. Chin Med J (Engl). 2015;128(9):1238–1244.
- Weeraratne SD, Amani V, Neiss A, et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro-Oncology. 2011;13(2):165–175.10.1093/neuonc/noq179
- Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–7576.10.1158/0008-5472.CAN-09-0529
- Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299(1):E110–E116.10.1152/ajpendo.00192.2010
- Beier D, Hau P, Proescholdt M, et al. CD133+ and CD133- glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010–4015.10.1158/0008-5472.CAN-06-4180
- Jin X, Jin X, Jung J-E, et al. Cell surface nestin is a biomarker for glioma stem cells. Biochem Biophys Res Commun. 2013;433(4):496–501.10.1016/j.bbrc.2013.03.021
- Soda Y, Hanahan D. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci USA. 2011;108(11):4274.10.1073/pnas.1016030108
- Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4(8):e6816.10.1371/journal.pone.0006816
- Chen YS, Chen ZP. Vasculogenic mimicry: a novel target for glioma therapy. Chin J Cancer. 2014;33(2):74–79.10.5732/cjc.012.10292