ABSTRACT
Here, we show that semiconductor-based sequencing technology can be used to map mammalian replication domains, chromosomal units with similar DNA replication timing. Replicating DNA purified from mammalian cells was successfully sequenced by the Ion Torrent platform. The resultant replication domain map of mouse embryonic stem cells is comparable to those obtained by the conventional microarray-based method.
DNA replication in mammalian cells is regulated as Mb-sized large chromosomal units called replication domains in which activation of multiple replication origins occurs in a relatively synchronous manner [Citation1,Citation2]. Genomic regions replicated in the first and second half of the S phase are called early and late replication domains, respectively. Early replication domains are basically characterized as gene-rich euchromatins, while late replication domains are relatively gene-poor and enriched with inactive chromatin marks. An intriguing fact revealed by the Hi-C experiment that early and late replication domains are largely consistent with A and B compartments of self-interacting chromatin domains suggests that replication domain organization reflects three-dimensional higher-order chromatin folding patterns in the nucleus [Citation3,Citation4]. A method to map replication domains at the genome-wide level was reported by David Gilbert and colleagues nearly ten years ago [Citation5]. In brief, early and late S phase fractions of BrdU-labeled cells are collected by flow cytometry. BrdU-labeled replicating DNA is enriched by immunoprecipitation with anti-BrdU antibody from each fraction. Isolated early and late replicating DNA are amplified and differentially labeled with fluorescence (e.g. Cy3 and Cy5) before hybridizing to the comparative genomic hybridization (CGH) microarray platform. By plotting the signal ratio of early and late replicating DNA for each CGH probe against the genomic position, periodic distribution of early and late replication domains along chromosome arms is detected. Initial studies to map replication domains rely mainly on commercial oligonucleotide microarrays, some of which are already discontinued. However, recent advances in next generation sequencing (NGS) techniques provide additional platforms for this type of genome-wide analysis. In the present study, we examined if ion torrent semiconductor sequencing platform, which employs pH measurement-based detection of nucleotide sequences, is applicable to map replication domains genome-wide in mammalian cells.
46C mouse embryonic stem (ES) cells were used because the CGH microarray-based replication domain map of this cell line is already available for comparison [Citation5,Citation6]. Experimental parts except whole-genome amplification (WGA) are essentially identical to the conventional method [Citation7]. Cells were labeled with 50 μM BrdU for 2 h, washed twice with ice-cold PBS, trypsinized, and then fixed in 75% ethanol. These cells were resuspended in PBS containing 1% FBS, stained with propidium iodide (20 μg/ml) for 30 min in the presence of RNase A (250 μg/ml), and then were sorted into early and late S phase fractions by FACSCalibur flow cytometer (BD Biosciences) (). After phenol-chloroform extraction of DNA from 10,000 cells, sonication was performed to cut the DNA into an average size of about 100–600 bp. Then, immunoprecipitation with anti-BrdU mouse monoclonal antibody (BD Biosciences, #555,627) was performed in each fraction to enrich BrdU-substituted replicating DNA. Isolated early and late replicating DNA was amplified individually by WGA. We modified the WGA step for NGS library preparation. Generally, DNA after WGA carries universal adaptor sequences at the ends, which negatively affects mappability of sequence reads to the reference genome. To overcome this problem, we used universal adaptor sequences that can be removed by restriction enzyme digestion after WGA (Figure S1). The resulting DNA molecules were used to construct libraries with Ion Plus Core Module for AB Library Builder System (Thermo Fisher Scientific, #4,477,683) and Ion Xpress Barcode Adapters 1–16 Kit (Thermo Fisher Scientific, #4,477,683). Sequencing was performed on the Ion PI™ Chip kit v3 chip using Ion Proton semiconductor sequencer (Thermo Fisher Scientific). Reads were mapped to the mm10 mouse reference genome using Bowtie2[Citation8] and reads with MAPQ< 20 were omitted from downstream analysis (). Data analyses were performed using SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and R/Bioconductor (http://www.r-project.org; http://www.bioconductor.org). Numbers of total reads in each sample were normalized per million reads in SeqMonk. The sequence reads ratio between early and late S phase samples was determined in 5-Kb windows and loess-smoothed over a 300-Kb window size in R/Bioconductor. The smoothed dataset was used to generate a replication domain map. Data were deposited in Gene Expression Omnibus (GEO) database under the accession number GSE115847.
Figure 1. Mapping replication domains using the ion torrent semiconductor sequencing platform.
(a) Flow chart of sample preparation for genome-wide replication domain mapping. (b) Summary of Ion Proton sequencing run for early and late replicating DNA samples. Q20 represents per-base quality score in Phred scale (only 1 alignment error is predicted to occur every 100 bases). (c) Replication domain maps of the 46C mouse ES cell (chromosome 16) from sequencing by Ion Proton and the previous CGH microarray-based data set. Loess-smoothed plots of log2 (early/late) ratio determined at each 5-Kb genomic bin are shown. Data sets were quantile-normalized for comparison. A merged plot on the bottom shows remarkable similarity between two data sets. (d) Expanded plots for an exemplary ES cell-specific replication domain (chr16: 48.5 Mb). The domain indicated by the arrowhead is early replicating in ES cells (46C, D3, and TT2), while the same domain is late replicating in mouse embryonic fibroblast cells (MEF) and myoblast cells (Myo). Data sets are averaged into 200-Kb fixed windows for correlation analysis. CGH microarray-based data sets from various cell types were downloaded from Replication Domain (http://www.replicationdomain.org). (e) Correlation analysis of replication domain maps from various cell types (Pearson’s R values).
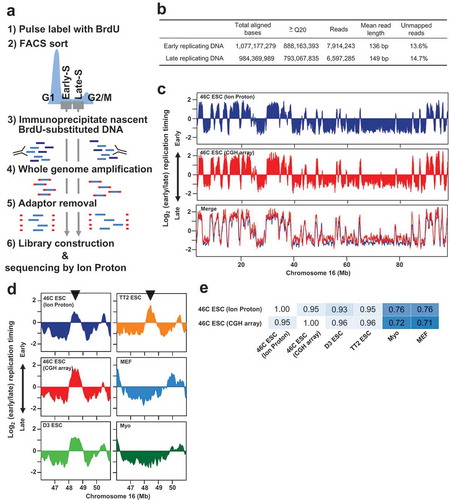
We found that obtained replication domain map of 46C ES cells is almost identical to the previous CGH microarray-based replication domain map from the same cell line. Mb-sized replication domains either early or late are highly conserved between two maps (, R = 0.95). A representative ES-cell specific early replicating domain identified in the previous study[Citation4,Citation5] is also detected in the Ion Proton-based replication domain map (). The Ion Proton-based map of 46C ES cells shows a higher genome-wide correlation with CGH microarray-based maps of several ES cell lines than differentiated cell types (), which further proves the correct mapping of cell-type specific replication domain organization.
Collectively, our study demonstrated the utility of Ion Torrent semiconductor-based sequencing platform for mapping mammalian replication domains. We used the mouse ES cell line as an example, though any species that has a sequenced genome can be analyzed using the same protocol described here. Very recently, Illumina sequencing platform, which employs fluorescence-based sequencing, was also shown to be applicable for replication domain mapping [Citation9]. In addition to the differences in the sequencing platforms, our protocol differs from the one used for Illumina sequencing in that BrdU-labeled DNA is immunoprecipitated before library construction and WGA is employed to obtain sufficient DNA for library construction. We confirmed that both protocols generate similar replication domain maps from 46C ES cells (Figure S2). These modified protocols for NGS will greatly facilitate the analysis of replication domain regulation and related chromatin dynamics.
Supplemental_figure_2.pdf
Download PDF (118.8 KB)Supplemental_figure_1.pdf
Download PDF (27.5 KB)Acknowledgments
We thank the Center for Molecular Biology and Genetics, Mie University, for sequencing service.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Rivera-Mulia JC, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect-part III. Curr Opin Cell Biol. 2016;40:168–178.
- Takebayashi SI, Ogata M, Okumura K. Anatomy of mammalian replication domains. Genes (Basel). 2017;8:110.
- Ryba T, Hiratani I, Lu J, et al Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770.
- Takebayashi S, Dileep V, Ryba T, et al Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc Natl Acad Sci USA. 2012;109:12574–12579.
- Hiratani I, Ryba T, Itoh M, et al Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245.
- Ying QL, Stavridis M, Griffiths D, et al Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186.
- Ryba T, Battaglia D, Pope BD, et al Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc. 2011;6:870–895.
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359.
- Marchal C, Sasaki T, Vera D, et al Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat Protoc. 2018;13:819–839.
