ABSTRACT
Oxaliplatin (L-OHP) is one of the effective chemotherapeutic drugs for colorectal cancer (CRC). Further investigation into the molecular mechanism of chemoresistance could improve outcomes for patients with colorectal cancer. Recently, microRNAs have been reported as a key in drug resistance of tumors. In this study, we aimed to investigate the effects of miR-153-5p in L-OHP-resistant CRC cells, and its underlying mechanism. Downregulation of miR-153-5p was observed in CRC cells, while upregulation of miR-153-5p enhances the chemosensitivity of CRC/L-OHP cells. The autophagy of CRC/L-OHP cells was markedly increased after exposure to L-OHP but abolished by the upregulation of miR-153-5p. Dual-luciferase reporter assays validated that Bcl-2 was a direct target of miR-153-5p. In conclusion, our data suggested that miR-153-5p was a mediator of cisplatin resistance in colorectal cancer by affecting Bcl-2-mediated autophagy, indicating a new therapeutic target for CRC treatment.
GRAPHICAL ABSTRACT

MiR-153-5p was a mediator of cisplatin resistance in colorectal cancer by affecting Bcl-2-mediated autophagy
Colorectal cancer (CRC) is the most common intestinal and gastric malignant tumor type, which is responsible for ~10% of cancer cases worldwide and is also a common cause of cancer-associated mortality [Citation1,Citation2]. Recently, the incidence of CRC has increased significantly, and 50% of patients eventually succumb to the disease because of metastatic spread of cancer [Citation3]. Surgery combining chemotherapy is the standard treatment for CRC; however, in recent decades, CRC has become resistant to several chemotherapeutic drugs such as oxaliplatin, which is one of the first-line chemotherapy treatments of CRC [Citation4].
MicroRNAs (MiRNAs) are a class of small non-coding RNAs, 18–24 nucleotide RNAs that regulate expression of a wide variety of genes by targeting their mRNAs, which leads to mRNA degradation or inhibition of protein translation [Citation5,Citation6]. In recent years, miRNAs are reported as a potential therapeutic strategy for cancer treatment even for the chemotherapeutic drugs resistant including CRC [Citation7]. MiRNA also implicated in tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers [Citation8]. As a member of the microRNA family, miR-153 has been shown to suppress or promote cancer cell proliferation, invasion, and migration [Citation9,Citation10]. It had been reported that miR-153 supports CRC progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance at the same time [Citation11]. A recent study reported that the expression of miR-153-5p was obviously negatively associated with Bcl-2 in radioresistance of human glioma [Citation12]. However, the biological importance of miR-153-5p in CRC and oxaliplatin-resistance remains still largely unknown.
Autophagy is a self-degradation process, which is essential for cell survival, differentiation, development, and homeostasis [Citation13]. It plays an adaptive role in protecting organisms against diverse pathologies. Abnormal autophagy is closely associated with many diseases, including tumors and drug-resistance. Studies have demonstrated that autophagy promotes cancer cell survival under specific conditions, and inhibits chemosensitivity of cancer cells [Citation14,Citation15]. It has been reported that circHIPK3 promotes L-OHP resistance in CRC by sponging miR-637, which is dependent on the inhibition of autophagy-related cell death via STAT3/Bcl-2/beclin1 signaling pathway [Citation16]. Zou Y et al. have reported that miR-153 regulates apoptosis and autophagy of cardiomyocytes by targeting Mcl-1 [Citation17]. We found that Bcl-2 has a predicted targeting binding site of miR-153-5p (http://www.targetscan.org). The present study aimed to determine the roles of Bcl-2 mediated autophagy and miR-153-5p, and their underlying mechanisms in oxaliplatin-induced colorectal cancer cells.
Materials and methods
Cell culture
SW480, HCT116 cell lines used in our study were purchased from American type culture collection (Rockefeller, MD, USA). SW480/L-OHP, HCT116/L-OHP refers to L-OHP (Hengrui Medicine, Lianyungang, Jiangsu, China) resistant cell lines, respectively, which were constructed by our lab. At first, we prepared a cell suspension at the concentration of about 1 × 105 cells/mL, and L-OHP was added into cell suspension and diluted L-OHP with cell suspension to the concentration of 4 μmol/mL. And after treatment over 48 h by L-OHP, we discarded the L-OHP and added fresh medium instead. Cells were passaged regularly and increased concentration of L-OHP to 5 μmol/mL, cells were also treated under L-OHP over 48 h, replaced the L-OHP with fresh medium. We repeated this treatment until the concentration of L-OHP was increased to 15 μmol/mL in order to collect the stable L-OHP resistant SW480 and HCT116 cell lines. Dulbecco’s Modified Eagle Medium (DMEM) (GE Healthcare, Little Chalfont, Buckinghamshire, UK) with 10% fetal bovine serum (Thermo Fisher Scientific, Boston, Massachusetts, USA) and 100 U/mL penicillin, and 100 mg/mL streptomycin (Thermo Fisher Scientific) were used for cell line culture. Both SW480 and SW480/L-OHP were cultured at 37°C in 5% CO2. L-OHP was purchased from Hengrui Medicine (Lianyungang, Jiangsu, China).
Plasmid transfection
MiR-153-5p mimic was obtained from GeneScript (Nanjing, China). PcDNA-BCL2 was constructed by our laboratory. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used for cell transfection according to the protocol provided by Invitrogen.
RNA extraction and qRT-PCR
RNAiso plus (Takara, Shiga, Japan) were used for RNA extraction. MMLV reverse transcription kit (Promega, Madison, WI, USA) was used for cDNA synthesis. SYBR Premix Ex Taq kit (Takara) was used for qRT-PCR and β-actin was used as an internal control. All these experiments were conducted according to the production manuals. Primers used in qRT-PCR were listed as follows:
MiR-153-5p FP: CGGTGTCATTTTTGTGACGT
MiR-153-5p RP: CAATGATCACTTTTGTGACT
Bcl-2 FP: TCCTCCCAGCCCGGGCACAC
Bcl-2 RP: CACCGGGCTGAGCGCAGGCC
p62 FP: GCCTGTGCTGGACCAGCTGG
p62 RP: GTCCTGGGAAGGAGCTATGG
LC3 FP: CGAACAAAGAGTAGAAGATG
LC3 RP: ACCAACAGGAAGAAGGCCTG
β-actin FP: CTCCATCCTGGCCTCGCTGT
β-actin RP: GCTGTCACCTTCACCGTTCC
Cell counting kit-8 assay
Cell viability of CRC cell lines was measured by Cell counting kit-8 assay. CCK-8 was purchased from Thermo Fisher Scientific. Cells were plated onto 96-well plates (4 × 104 cells/well), each well was supplemented with 10 μL of CCK-8 solutions for 1 h at 37 °C. At last, we read the absorbance value of each well at 450 nm by a microplate reader (Thermo Fisher).
Flow cytometry analysis
Flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and Annexin V-FITC Apoptosis Detection kit (BD Biosciences) was used to determine cell apoptosis of CRC cell lines. Cells were first washed by PBS three times, and then diluted into single-cell suspension. After re-suspending with the binding buffer, cells were stainedwith 10 μL Annexin V-fluorescein isothiocyanate (FITC), and 5 μL propidium iodide (PI) for 10 min, in the dark. At last, we conducted fluorescence-activated cell sorting (FACS).
Western blotting analysis
Briefly, total protein from CRC cell lines were extracted by RIPA buffer (Beyotime Institute of Biotechnology, Haimen, P.R. China) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% resolving gel), and then transferred the gel to a PVDF membrane (GE Healthcare). Corresponding appropriate primary antibody was used to incubate the membranes, after incubation by secondary antibody, the results were detected by HRP-mediated chemiluminescence (Millipore, Billerica, USA). ECL reagents were used to detect the protein bands, and the data were accessed by ImageJ software (National Institutes of Health, MD, USA). Primary antibody used in this study was listed as follows: anti-Bcl-2 (Cell Signaling Technology), anti-p62 (Cell Signaling Technology), anti-LC3 I and II (Abcam), anti-beclin (Cell Signaling Technology), and anti-β-actin (Abcam). Secondary antibody were purchased from Beyotime Institute of Biotechnology.
Immunofluorescence staining assay
We seeded the cells in culture dishes. And then we fixed cells with 4% paraformaldehyde for 20 min when confluence reached 60–70% and washed cells with PBS for 3 times. Triton X-100 (0.2%) was used for permeabilizing the cells, and after permeabilized, 5% BSA was added into the dish immediately and incubated for 30 min at room temperature. Primary antibodies were used to incubate the cells at 4°C overnight, and then secondary antibodies were used to incubate the cells. At last, we stained the nuclear with 4,6-diamino-2-phenylindole (DAPI, Solarbio, Beijing, China). Washed off DAPI with PBS and obtained images with a fluorescence microscope.
Luciferase assay
We specifically cloned Plasmids containing wild-type Bcl-2 or Bcl-2 mutant 3ʹUTR into the pMIR-report-luciferase vector (Invitrogen). Dual-Luciferase Reporter Assay System (Promega) was used to perform luciferase activity assays following the production manuals.
Statistical analysis
Data were expressed as the mean ± standard error of the mean (SEM). One-way ANOVA was performed in multigroup comparison. A value of p < 0.05 was considered to indicate a statistically significant difference.
Results
MiR-153-5p downregulated in CRC/L-OHP cells and regulate chemoresistance
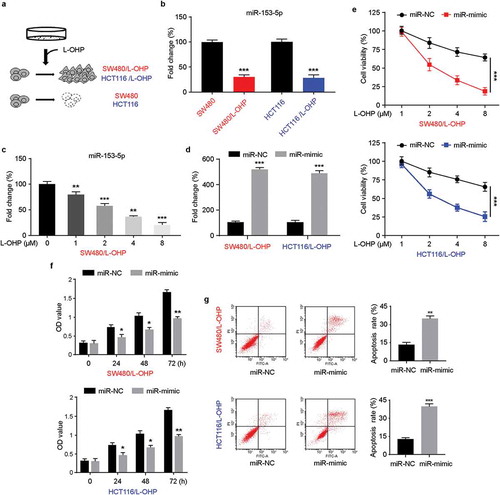
L-OHP-resistant SW480/L-OHP, HCT116/L-OHP cell lines were established through L-OHP treatment ()). The mRNA levels of miR-153-5p were measured by qRT-PCR, the results indicated that miR-153-5p was significantly downregulated in L-OHP-resistant CRC cell lines ()). Seventy-two hours after L-OHP treatment, the expression of miR-153-5p was downregulated with a dose-dependent manner in L-OHP-resistant SW480 cell lines ()). To explore the role of miR-153-p, we transfected miR-153-5p mimic into L-OHP-resistant CRC cells ()). The upregulation of miR-153-5p effectively inhibited the cell viability and cell growth of CRC/L-OHP under L-OHP treatment (,). Flow cytometry results showed that overexpressed miR-153-5p induced increased cell apoptosis. Altogether, our results suggested that miR-153-5p played aregulatory role in L-OHP resistant in CRC cells.
L-OHP could induce cells autophagy in CRC/L-OHP
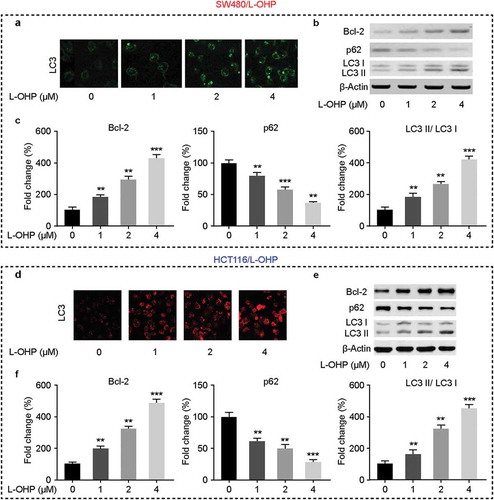
Autophagy was implicated in the chemoresistance of cancer cells [Citation12]. Next, we validated the effects of L-OHP on cells autophagy in CRC/L-OHP cells 72 h after the treatment. We performed immunofluorescence and western blot to determine the key factors in the process of cells autophagy. Immunofluorescence staining assay suggested that in CRC/L-OHP cell lines under the treatment of L-OHP, LC3 was upregulated with the increasing concentration of L-OHP (,). Western blot results showed that, under the treatment of L-OHP, the protein levels of BCL2 were upregulated, and p62 were downregulated with the increasing concentration of L-OHP (–) in both SW480/L-OHP and HCT116/L-OHP cell lines. These data indicated that the treatment of L-OHP induced cells autophagy.
Bcl-2 was a direct target of miR-153-5p
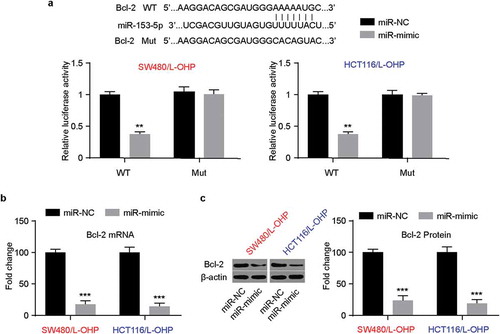
In order to search for the potential targets of miR-153-5p, we conducted bioinformatic analysis with TargetScanHuman 7.2, and there was a potential target of miR-153-5p on Bcl-2 ()). The results of luciferase reporter assay system showed that in Bcl-2 wild-type CRC/L-OHP cell lines, the relative luciferase activity was downregulated significantly. However, in Bcl-2 mutant CRC/L-OHP, there was no notable variation. Furthermore, qRT-PCR and western blot were conducted. As is shown in ,, the mRNA levels and protein levels of Bcl-2 downregulated significantly in miR-153-5p overexpressed CRC/L-OHP cell lines. These data revealed that Bcl-2 was the potential targets of miR-153-5p, and miR-153-5p could downregulate Bcl-2.
MiR-153-5p could inhibit cells autophagy through down-regulating the expression of Bcl-2
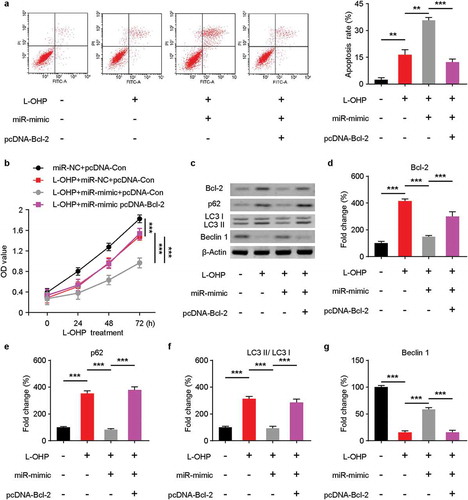
To further explore whether miR-153-5p could target Bcl-2 and enhance the sensitivity to L-OHP and autophagy in SW480 cell lines. The SW480 cells transfected with miR-153-5p-mimic or/and Bcl-2 plasmid were treated with 8 μM L-OHP for 72 h. As is shown in ), the apoptosis rate of SW480 cell lines was increased after L-OHP treatment and miR-153-5p mimic overexpression but decreased after Bcl-2 and miR-153-5p overexpressed. In the CCK8 assay, co-transfected miR-153-5p mimic and Bcl-2 increased SW480 cells survival rate significantly, compared to transfected miR-153-5p mimic singly ()). Then, we performed western blot to check the protein levels of autophagy-related proteins ()). The protein levels of Bcl-2, p62, LC3 (–, respectively) decreased after miR-153-5p overexpression in SW480 cells treated with L-OHP, whereas they increased after we overexpressed Bcl-2 and miR-153-5p. And Beclin 1 was upregulated after miR-153-5p mimic transfection in SW480 cells under the treatment of L-OHP, while the tendency reversed upon Bcl-2 upregulation. Taken these findings together, they indicated that miR-153-5p enhanced sensitivity of SW480 cells to L-OHP and autophagy through targeting Bcl-2.
Discussion
Numerous pieces of evidence had revealed that aberrant miRNA expression plays a functional role in CRC initiation and progression [Citation18]. Oxaliplatin is one of the most effective chemotherapeutic agents in the management of CRC. However, nowadays, oxaliplatin chemoresistance has become a serious problem in tumor treatment including CRC. It is urgent to explore the potential mechanisms of chemoresistance and take strategies to increase the chemosensitivity of CRC cells.
Here, we aimed to Figure out the role of miR-153-5p and Bcl-2 mediated autophagy in oxaliplatin chemoresistance on CRC. In this study, a marked decrease in miR-153-5p expression was observed in oxaliplatin-resistance SW480/L-OHP cells. Moreover, miR-153-5p-mimic induced higher apoptosis rates and suppressed cell viability compared with miR-153-5p-NC with or without oxaliplatin treatment. It had been reported that miR-153 regulates the apoptosis and autophagy of cardiomyocytes by targeting Mcl-117, suggested that a potential interaction between miR-153-5p and autophagy inregulating oxaliplatin resistance in CRC.
Accumulating evidence indicates that autophagy, a cellular catabolic process, which occurs in response to stress conditions and specific environmental factors, including chemotherapeutic agents, pathogen infection, and nutrient deprivation, can promote cell survival [Citation19,Citation20]. Autophagosome, a double-membrane-bound vacuole that fuses with the lysosome and engulfs cellular material targeted for degradation, resulting in the degradation of its contents [Citation21]. Beclin-1, an interacting partner of the antiapoptotic protein Bcl-2, behaves as an autophagy-related protein which plays a role in the formation of autophagosomes. It also involves the cytosolic form of microtubule-associated protein 1 light chain 3 (LC3-I). LC3-I, a cytosolic form of LC3, is conjugated to phosphatidylethanolamine to form LC3-II, which is localized to the autophagosomal membrane and degraded after fusion of autophagosomes to lysosomes [Citation22]. Thus, the LC3-II/LC3-I ratio is a measure of autophagic activity. Autophagy has been reported to promote resistance of hepatocellular carcinoma cells to oxaliplatin, but the role in CRC is not clear. In our study, both immunofluorescence and western blotting assays showed that the level of Bcl-2 and LC3-II/LC3-I in SW480/L-OHP cells was increased after oxaliplatin treatment, while p62 decreased, indicating the promoting role of autophagy in the resistance of CRC to oxaliplatin.
Bcl-2 is a potential target of miR-153-5p according to online prediction software and dual-luciferase assay. However, it is still unclear whether miR-153-5p regulates the chemosensitivity of CRC cells to oxaliplatin by targeting Bcl-2. To determine the functional mechanism of the effects of miR-153-5p on autophagy, the protein expression level of Bcl-2 was detected in SW480/L-OHP cells treated with oxaliplatin. Results showed that both mRNA and protein expression level of Bcl-2 was significantly inhibited when miR-153-5p was overexpressed. Moreover, cell viability increased and the apoptosis rate decreased after Bcl-2 and miR-153-5p overexpression. The protein levels of Bcl-2, p62, LC3-II/LC3-I decreased after miR-153-5p overexpression in SW480 cells treated with L-OHP, and Beclin 1 was upregulated. However, the tendency was recovered by miR-153-5p-mimic and pcDNA-Bcl-2 co-transfection.
Taken together our results demonstrate that miR-153-5p affects oxaliplatin-tolerance in CRC. Specifically, it promotes sensitivity of CRC cells to oxaliplatin while targeting Bcl-2 mediated autophagy, suggesting a new therapeutic target for overcoming drug resistance in CRC.
Author contributions
Y.X-Z designed the research. H.Y, Z.L, and T.F performed experiments, analyzed the data, and wrote the paper. W.L-F and L.D-H performed the illustrations of the data. All authors have read and approved the final manuscript.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Toru A, Kosuke K, Koji O, et al. Clinical impact of tumor location on the colon cancer survival and recurrence: analyses of pooled data from three large phase III randomized clinical trials. Cancer Med. 2017;6(11):2523–2530.
- Jacques F, Hai-Rim S, Freddie B, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917.
- Sarah K, Ilana N, Shiran S, et al. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347(1):15–21.
- Chengyao Z, Jiawu W, Haitao G, et al. Capecitabine plus oxaliplatin compared with 5-fluorouracil plus oxaliplatin in metastatic colorectal cancer: meta-analysis of randomized controlled trials. Oncol Lett. 2012;3(4):831–838.
- Jing-Jia Y, Jiang C. MicroRNAs in colorectal cancer as markers and targets: recent advances. World J Gastroenterol. 2014;20(15):4288–4299.
- Grady William M, Pritchard Colin C. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol. 2014;42(1):124–139.
- Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17(8):523–531.
- Xu Q, Jing Y, Yu Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12(9):1385–1394.
- Jun Z, Ming X, Ying S, et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol Rep. 2015;34(1):111–120.
- Wenkang L, Yan S, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29.
- Lei Z, Karen P, Veronika J, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–6447.
- Deyu S, Yi M, Haozhe P. MicroRNA-153-3p enhances cell radiosensitivity by targeting BCL2 in human glioma. Biol Res. 2018;51(1):56.
- Lai K, Killingsworth MC, Lee CS. The significance of autophagy in colorectal cancer pathogenesis and implications for therapy. J Clin Pathol. 2014;67(10):854–858.
- Ankit K, Kumar SU, Anurag C. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7(12):1535–1542.
- Feifei L, Donglei L, Yang Y, et al. Effect of autophagy inhibition on chemotherapy-induced apoptosis in A549 lung cancer cells. Oncol Lett. 2013;5(4):1261–1265.
- Yanli Z, Chen L, Xinfeng L, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48(undefined):277–288.
- Yuhai Z, Wenting L, Zhang J, et al. miR-153 regulates apoptosis and autophagy of cardiomyocytes by targeting Mcl-1. Mol Med Rep. 2016;14(1):1033–1039.
- Cowland Jack B, Christoffer H, Kirsten G. MicroRNAs and cancer. APMIS. 2007;115(10):1090–1106.
- Naiara S-C, Mancias Joseph D, Kimmelman Alec C. The role of autophagy in cancer. Annu Rev Cancer Biol. 2017;1(undefined):19–39.
- Lum Julian J, Bauer Daniel E, Mei K, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120(2):237–248.
- Lamb Christopher A, Tamotsu Y, Tooze Sharon A. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774.
- Isei T, Takashi U, Eiki K. LC3 and autophagy. Methods Mol Biol. 2008;445(undefined):77–88.