ABSTRACT
KLF9 is reported to promote adipocyte differentiation in 3T3-L1 cells and pigs. However, the roles of KLF9 in adipocytes differentiation of goat remain unknown. In this study, the expression profiles of KLF9 were different between subcutaneous and intramuscular preadipocytes of goat during differentiation process. After silencing KLF9 gene, the lipid droplets were increased in both two types of adipocytes. In subcutaneous preadipocyte with silencing KLF9, the expressions of C/EBPβ, PPARγ, LPL, KLF1-2, KLF5, and KLF17 genes were up-regulated, while KLF12, KLF4, and KLF13 genes were down-regulated in expression level. In intramuscular preadipocyte, aP2, C/EBPα, KLF2-3, KLF5, and KLF7 gene were up-regulated, and Pref-1 gene was down-regulated. In addition, the binding sites of KLF9 existed in the promoters of aP2, C/EBPα, C/EBPβ, LPL and Pref-1. Taken together, KLF9 play a negative role in the differentiation of both intramuscular and subcutaneous preadipocytes in goats, but the functional mechanism may be different.
GRAPHICAL ABSTRACT

Knockdown KLF9 promotes differentiation of intramuscular and subcutaneous preadipocyte, however, the mechanism of action might be different.
Adipocyte differentiation is one of the important ways of fat deposition. During the complex biological process of adipocyte differentiation, many transcription factors play important roles, such as Peroxisome proliferator-activated receptors (PPARs), CCAAT enhancer-binding proteins (C/EBPs), Sterol regulatory element binding protein isoform 1(SREBP1), Kruppel like factors family (KLFs) and Preadipocyte factor-1 (Pref-1). Also, some metabolism of triglycerides genes are involved in this process, like lipoprotein lipase (LPL) and fatty acid binding protein (FABP4 or aP2) [Citation1–Citation3]. Due to intramuscular adipocytes are dispersed in muscle tissue, the deposition of intramuscular fat depends not only on the differentiation and proliferation of intramuscular adipocytes, but also on the rate of muscle growth along with its metabolic activity. Therefore, intramuscular adipocytes have a special pattern of proliferation and differentiation compared with other adipocytes [Citation4]. In pigs, it was demonstrated subcutaneous preadipocytes had higher proliferation and lipogenic potential than intramuscular preadipocytes [Citation5,Citation6]. Further studies have found that there were a large number of differentially expressed genes between intramuscular and subcutaneous adipocytes in several species, such as pigs and cattle [Citation7–Citation9]. However, the regulatory mechanisms of most genes on adipocyte differentiation of different parts of body are still unclear.
Krüppel like factors 9 (KLF9) belongs to the SP/KLF transcription factor family that is typically characterized by the presence of three highly conserved Cys2/His2 zinc finger structures [Citation10]. It was demonstrated that SP/KLF protein could regulate gene transcription by binding to the GC/GT cassette and CACCC elements in the promoter, as well as, the enhancer/silencer regions of the target gene [Citation11]. In recent years, studies have found that KLF9 plays an important role in the adipocytes differentiation. KLF9 promotes early differentiation of 3T3-L1 cells through activating the expression of C/EBPβ gene [Citation12]. However, the expression of KLF9 was significantly upregulated during the mid-differentiation. The expression of various lipogenesis-related genes, such as PPARγ, C/EBPα and aP2, were significantly reduced after KLF9 knockdown. Moreover, KLF9 may act to activate the PPARγ2 promoter by direct interaction with C/EBPα, or binding to the promoter region of PPARγ2 to promote adipogenesis [Citation13]. In addition, under the conditions of adipogenication, KLF9 can promote the accumulation of triglycerides in hepatocytes by regulating the expression of PPARγ [Citation14]. Peng and colleagues found that miR-429 inhibited the differentiation of subcutaneous progenitor cells in pigs by directly binding to the 3ʹUTR region of KLF9 [Citation15]. Previous studies indicate that KLF9 promotes adipogenic differentiation in 3T3-L1 cells and pigs. However, due to species specificity, the same gene may have different regulatory effects on different type cells or different part fat cells of body even in the same species [Citation16]. Is the KLF9 gene involved in goat adipocyte differentiation? If so, is there a difference in the mechanisms that regulate the adipocytes differentiation in different parts of body (subcutaneous preadipocytes and intramuscular preadipocytes)?
In this study, we screened the expression pattern of KLF9 during the differentiation of both subcutaneous and intramuscular preadipocytes in goat, and transfected siRNA-KLF9 into both subcutaneous and intramuscular preadipocytes of goat. Morphological methods were used to observe changes in cell lipid droplet aggregation after KLF9 knockdown. The qPCR technique was used to detect the expression changes of adipocyte differentiation marker genes and other members of KLFs after KLF9 knockdown. Finally, we determined the effects of KLF9 on the differentiation of both subcutaneous and intramuscular adipocytes in goats with the possible mechanisms.
Materials and methods
Isolation and culture of cells
Animal studies were approved by the Institutional Animal Care and Use Committee, Southwest Minzu University (permit number: 2011-3-2). The isolation and culture of goat intramuscular preadipocytes was performed as described assay by Xu et al. [Citation17]. The subcutaneous adipocytes were isolation by using Type I collagenase. The longissimus dorsi and subcutaneous fat were excised from 7-day-old goats and minced. Collagenase TypeⅡ(37°C, 1.5 h) (Sigma) and collagenase typeⅠ(37°C, 1 h) (Sigma) were used to digest the pellets of longissimus dorsi and subcutaneous fat, respectively. Enzymatic digestion was terminated by the same volume of DMEM/F12 (Hyclone) supplemented with 10% FBS (Gemin). The suspension was filtered through a 75 µm nylon cell strainer, and then centrifuged at 750 rpm/min for 5 min. After disposing of the red blood cell lysed solution, the suspension was centrifuged at 750 rpm/min for 5 min again. The pre-adipocytes were re-suspended in DMEM/F12 supplemented with 10% FBS, and diluted to a final concentration of 106 cells/mL. These cells were cultured at 37°C under a humidified atmosphere containing 5% CO2.
Design of siRNA
According to the sequence of goat KLF9 gene obtained in the previous investigation [Citation17], siRNA targeting KLF9 gene was designed by the online program: RNAi Designer (http://rnaidesigner.thermofisher.com/rnaiexpress/) and synthesized by Invitrogen. The corresponding Negative Control is provided by Invitrogen (). The siRNA was centrifuged at 12,000 rpm/min for 10 min and dissolved in 125 μL of DEPC water to a final concentration (20 μmol/L).
Table 1. Sequences of siRNAs for goat KLF9.
Passage and induce of cells
When the cell confluence reaches 80%, we discarded the medium and added appropriate amount of trypsin, and the cells were digested at 37°C for 1 min. And then, the cells were harvested in a centrifuge tube, followed by a centrifuge at 800 rpm/min for 3 min to obtain cell pellet. The collected cells were re-suspended for subsequent experiments. The cells, passaged to the third generation, were used for experimental treatment. The cells were coaxed into differentiating using oleic acid induction solution (50 μmol/L), and harvested after induction for 0, 2, 4, and 6 d. These cell samples were used to construct temporal expression profile.
siRNA transfection
When the cell confluence reaches 80%, the cells were cultured for 4 h in Opti MEM medium before transfection to lead to serum starvation. The mixture was incubated for 5 min at room temperature with Opti MEM: RNAi MAX Reagent: siRNA = 3:2:25, the final concentrations of the mixture of siRNA was 80 nmol/L. After 6 h of transfection, cells were induced for differentiation with oleic acid induction solution (50 μmol/L). At differentiation for 48 h, the cells were washed 3 times with PBS and were collected for further analysis.
Oil red O and Bodipy staining
The Oil red O staining was carried out following the protocol described by Xu and colleagues [Citation18]. The Bodipy staining was performed according to the method described by Cai et al. [Citation19]. Briefly, the cells were fixed with 4% formaldehyde for 15 min and washed with PBS. Then the cells were stained using Oil red O solution and Bodipy solution (away from light) for 20 min, respectively. After staining, the cells were pictured using an Olympus TH4-200 microscope (Tokyo, Japan) for magnification views. Oil Red O dye was extracted with 100% isopropanol, and Oil red signal quantified by measuring the absorbance at 490 nm (OD 490) to determine the extent of differentiation.
Total RNA extraction, cDNA synthesis and real-time PCR (qPCR)
Total RNA was extracted by TRIzol regent and reversely transcribed into cDNA. The cDNA was diluted 10-fold for qPCR detection. The qPCR protocol was used to detect the silencing effect of KLF9 gene, the expression level of marker genes for differentiation (aP2, C/EBPα, C/EBPβ, LPL, PPARγ, Pref-1 and SREBP1) and the expression changes of other members of KLFs. The UXT gene was used as an internal reference. The information of qPCR primers is shown in .
Table 2. Primers for quantitative real-time PCR (qPCR).
Data analysis
The data was homogenized by 2−ΔΔCt method, and recorded into SPSS 18.0 software system (SPSS Inc., Chicago, IL, USA) for variance analysis by T-test. The data was expressed as “Mean ± standard deviation (SD)”. ** indicates extremely significant difference (P < 0.01), and * indicates a significant difference (P < 0.05).
Results
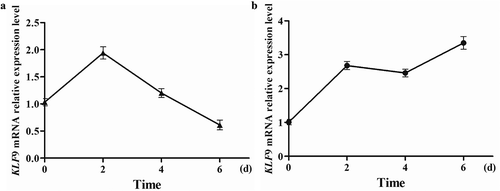
Relative expression level of KLF9 during the differentiation of intramuscular and subcutaneous preadipocytes in goat
Intramuscular and subcutaneous preadipocytes of goat were cultured in vitro. The cells of passage 3 were harvested at days 0, 2, 4, and 6 after induction of differentiation. The relative expression level of KLF9 was detected during different stages of cells. Our previous results showed that the relative expression of KLF9 increased during differentiation of intramuscular preadipocytes from 0 to 2 d and continued to decrease from 2 to 6 d () [Citation20]. But the expression level of KLF9 increased rapidly in the differentiation of subcutaneous preadipocytes from 0 to 2 d, while the relative expression of KLF9 increased slightly in 2–4 d and increased in 4–6 d during the differentiation process ().
KLF9 inhibits the differentiation of intramuscular and subcutaneous preadipocytes in goat
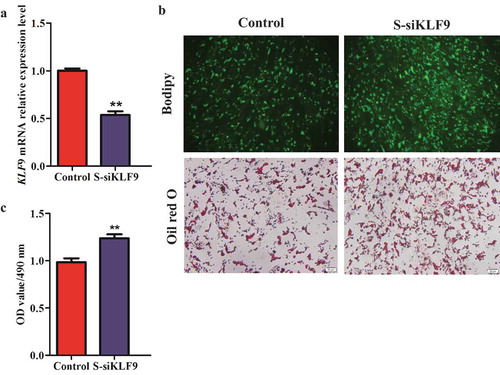
To explore the role of KLF9 gene in the differentiation regulation of intramuscular and subcutaneous preadipocytes in goat, we used siRNA to silence the expression of KLF9 gene in intramuscular and subcutaneous preadipocytes of goats. The relative mRNA expression of KLF9 was decreased by ~68.31% and ~46.50% compared with the control (P < 0.01), respectively, based on the use of siKLF9 ( and ). Knockdown of KLF9 promoted lipid droplet accumulation, as evidenced by oil red O and Bodipy staining. At the same time, the OD value of OD490 nm of the siKLF9 group was significantly increased in the intramuscular and subcutaneous adipocytes (P < 0.01), which was 1.09 (c) times and 1.21 times versus the Control group, respectively (c).
Figure 2. The silencing efficiency analysis of KLF9 gene in intramuscular preadipocytes of goats. (a) The silencing efficiency of KLF9 was detected in mRNA level, n = 6. (b) Morphological observation of Oil red O staining and Bodipy staining. (c) The OD490 nm value was detected by Oil red O staining, n = 6. * extremely significant difference (P < 0.01).
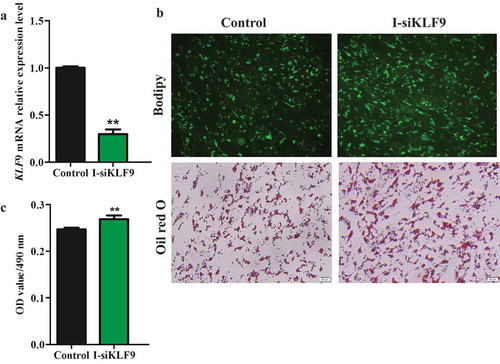
Figure 3. The silencing efficiency analysis of KLF9 gene in subcutaneous preadipocytes of goats. (a) The interference efficiency of KLF9 was detected in mRNA level, n = 6. (b) Morphological observation of Oil red O staining and Bodipy staining. (c) The OD490 nm value was detected by Oil red O staining, n = 6. ** extremely significant difference (P < 0.01).
Effect of KLF9 on the expression of adipocyte differentiation marker genes
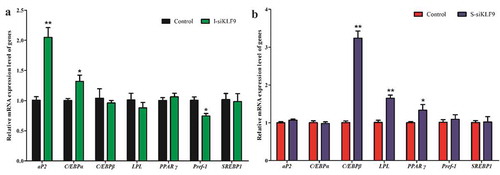
To further determine the role of KLF9 gene in the preadipocytes differentiation of goats, we detected the expression of differentiation marker genes aP2, C/EBPα, C/EBPβ, LPL, PPARγ, Pref-1 and SREBP1 using qPCR technique. After silencing KLF9 in goat intramuscular preadipocytes, the expression levels of aP2 and C/EBPα were up-regulated by 2.04-fold (P < 0.01) and 1.32-fold (P < 0.05), respectively, and the expression level of Pref-1 was down-regulated by 26.00% (P < 0.01), while the expression changes of C/EBPβ, LPL, PPARγ? and SREBP1 genes were not significantly differentces (P > 0.05) (). However, silencing KLF9 in goat subcutaneous preadipocytes had no significant effect on the expression of aP2, C/EBPα and Pref-1 gene (P > 0.05), but up-regulated the relative expression of C/EBPβ (3.23 fold, P < 0.01), LPL (1.63 fold, P < 0.01) and PPARγ genes (1.40 fold, P < 0.05) (). Thus, it is speculated that KLF9 may play roles through differential mechanisms between subcutaneous and intramuscular adipocytes of goat.
Figure 4. The effect of silencing KLF9 on adipocyte differentiation related-maker genes in differentiation of goat preadipocytes. (a–b) The expression changes of aP2, C/EBPα, C/EBPβ, LPL, PPARγ? and Pref-1 in intramuscular adipocytes (a) and subcutaneous adipocytes (b). ** extremely significant difference (P < 0.01), * significant difference (P < 0.05), n = 6.
Effect of silencing KLF9 on the expression of other members of the KLF family
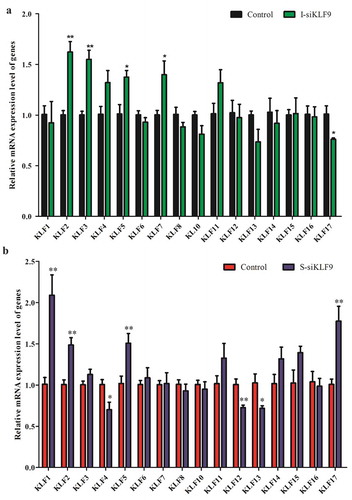
Based on the mutual regulation relationship among members of KLFs [Citation21,Citation22], we detected the relative expression changes of other members of KLFs after silencing KLF9 in goat preadipocytes. The results showed that the relative expression levels of KLF2, KLF3, KLF5 and KLF7 genes were up-regulated by 1.62 times (P < 0.01), 1.55 times (P < 0.01), 1.36 times (P < 0.05) and 1.39 times (P < 0.05) in intramuscular adipocytes, and the expression of KLF17 were down-regulated (P < 0.05). However, the expression of, KLF1, KLF2, KLF5 and KLF17 were up-regulated by 2.07-fold (P < 0.01), 1.47-fold (P < 0.01), 1.50-fold (P < 0.01) and 1.76-fold (P < 0.01) in subcutaneous adipocytes, respectively, while the expression of KLF4, KLF12 and KLF13 were down-regulated with varying degrees, by 30.44% (P < 0.05), 37.18% (P < 0.01) and 30.87% (P < 0.05), respectively ().
Figure 5. The effect of silencing KLF9 on the other members of KLFs during differentiation of goat preadipocytes. (a–b) The expression of other KLFs members was changed after silencing KLF9 in intramuscular adipocytes (a) and subcutaneous adipocytes (b). ** extremely significant difference (P < 0.01), * significant difference (P < 0.05), n = 6.
Comparison of the effects of KLF9 on the intramuscular and subcutaneous preadipocytes differentiation in goat
To comprehensively compare and analyze the differences in the effects of KLF9 on the differentiation of goat intramuscular and subcutaneous precursor adipocytes, Venny 2.1 program was used to cross the genes with significant changes in expression ().
Figure 6. Silencing KLF9 gene has discrepancy between subcutaneous adipocytes and intramuscular adipocytes. (a–b) Differentiation marker genes (a) and other members of KLFs (b) were differently regulated in intramuscular and subcutaneous adipocyte after silencing KLF9. (c) The KLF9 binding DNA motif. (d) The predicted KLF9 binding sites at the promoters of aP2, C/EBPα, C/EBPβ, LPL, PPARγ? and Pref-1.
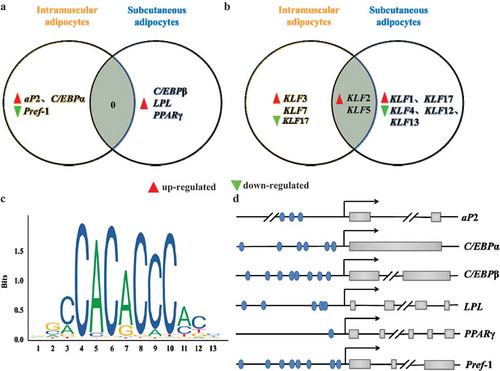
Silencing KLF9 can promote the differentiation of goat both intramuscular and subcutaneous adipocytes, but there is no intersection of the regulated differentiation marker genes (a). Differentiation is regulated in goat intramuscular adipocytes by up-regulating aP2 and C/EBPα along with down-regulated expression of Pref-1. It is speculated that silencing KLF9 promotes the differentiation of goat intramuscular fat cells by inhibiting lipid decomposition and promoting lipid production. However, in the subcutaneous adipocytes of goats, only the expression of C/EBPβ, LPL and PPARγ genes was up-regulated. Therefore, it was speculated that silencing KLF9 gene promoted the differentiation of goat subcutaneous adipocytes only by promoting lipid production. At the same time, KLF2 and KLF5 were up-regulated in both subcutaneous and intramuscular adipocytes after silencing KLF9 gene. However, the expression of KLF3 and KLF7 genes were also up-regulated in intramuscular fat cells, while KLF17 was inhibited. KLF1 and KLF17 were up-regulated in subcutaneous fat cells, but the expression of KLF4, KLF12 and KLF13 genes was down-regulated ().
In addition, online JASPAR (http://jaspar.genereg.net/) program was used to predict the DNA binding motif of KLF9 gene (). The predicted results are consistent with the binding of KLFs to the DNA region rich in GC and CACCC boxes, the binding sites of KLF9 gene are predicted in the promoter regions of aP2, C/EBPα, Pref-1, C/EBPβ, LPL and PPARγ genes. (d). Based on comprehensive comparative analysis and prediction results, it is speculated that KLF9 may have differential regulatory mechanisms in the differentiation of intramuscular and subcutaneous preadipocytes in goat. It was found that KLF9 may inhibit the differentiation of goat subcutaneous adipocytes by regulating the expression of C/EBPβ and LPL genes. However, it possibly inhibits the differentiation of goat intramuscular adipocytes by regulating the expression of aP2, C/EBPα and Pref-1 genes.
Discussion
Adipocyte differentiation is a complex process that responds to the hormonal sensitivity of various stimuli and coordinated changes in gene expression. Adipocyte differentiation requires sequential activation of transcription factors such as PPARγ, C/EBP, SREBP, and KLFs [Citation23]. The C/EBPβ and C/EBPδ are key transcription factors in the early stage of lipogenesis, and subsequent expression of C/EBPα and PPARγ genes is activated by these transcription factors [Citation24]. Various transcription factors, such as SREBPs and KLFs, play an important role in this process [Citation25]. Thus, all of these reveal the regulatory functions of transcription factors in adipogenesis, which is critical to elucidate the entire regulatory mechanism of adipogenesis. In this study, functional inhibition of KLF9 by siRNA indicated that KLF9 is a key transcription factor that differentially regulates adipocyte differentiation in different parts of goat body (intramuscular and subcutaneous preadipocytes). The expression level of KLF9 gene increased first and then decreased during the differentiation of goat intramuscular adipocytes. During the differentiation process of subcutaneous adipocytes, it increased at 0–2d. It is suggested that the KLF9 gene may play a similar or different regulatory pattern in two different types of preadipocytes.
The expression level of KLF9 gene during the differentiation of porcine subcutaneous preadipocytes increased 0–10 d, and then decreased, and the peak expression appeared on day 2 after differentiation [Citation15]. Sun et a. [Citation26] suggested that the expression level of KLF9 significantly increased during differentiation of intramuscular in chicken, and the expressional peak shown in day 4. Mori et al [Citation27] and Kimura et al [Citation12] found that the expression of KLF9 gene during the differentiation process of 3T3-L1 cells increased first and then decreased, but the expression peaks appeared on day 2 d and 1 h, respectively. However, Pei et al. [Citation13] pointed out that the expression level of this gene gradually increased with the differentiation in 3T3-L1 cells. We speculated that the differential expression pattern of KLF9 in the same adipocyte differentiation process may be caused by the difference between experimental conditions and differentiation time points.
To further investigate the biological role of KLF9 gene in goat adipocytes, this study silenced KLF9 in goat preadipocytes by RNAi technology. Oil red O and Bodipy staining showed that silencing KLF9 gene significantly promoted lipid accumulation in both subcutaneous and intramuscular preadipocytes. This goes against the results from 3T3-L1 adipocytes [Citation12,Citation13], suggesting that KLF9 has differential regulatory effects on adipocyte differentiation in different species. KLF13 promotes differentiation of porcine progenitor adipocytes, but has no significant effect on the differentiation of mouse 3T3-L1 adipocytes [Citation16], which is similar to our results.
The process of adipocyte differentiation is regulated by various transcription factors. These transcription factors are responsible for the expression of key proteins that induce the formation of mature adipocytes [Citation28]. In mammalian cells, PPARγ and C/EBPα are considered to be major regulators of lipogenesis [Citation29]. In addition, aP2, SREBP1, and LPL positively regulate adipocyte differentiation, while Pref-1 negatively regulates adipogenesis. They are key marker genes for adipocyte differentiation [Citation30]. Therefore, the above factors are used as criteria for judging differentiation. It was finally found that KLF9 affects cell differentiation by regulating the expression of aP2, C/EBPα, Pref-1 and C/EBPβ, as well as, LPL and PPARγ genes in goat intramuscular and subcutaneous adipocytes, respectively. Moreover, it was predicted by online programs that binding sites of KLF9 were present on the promoters of these altered differentiation marker genes. This demonstrates that KLF9 is involved in the differentiation of adipocytes in different parts of goat body by regulating different target genes. Previous studies have shown that KLF9 can promote adipogenic differentiation by binding to PPARγ or C/EBPβ [Citation12,Citation13]. This is similar to the genes regulated by KLF9 in goat subcutaneous adipocyte differentiation, but is not different from the genes regulated in goat intramuscular adipocytes differentiation. This indicates that the biology function of KLF9 has not only species specificity, but also cell specificity, which provides evidence for the specific differentiation pattern of intramuscular adipocytes [Citation4].
Members of KLFs are transcriptional activation or inhibitory factors that regulate mammalian adipocyte differentiation [Citation31], and there are cascaded regulatory properties among KLFs. For example, KLF1 and KLF3 are antagonistic in the regulation of erythropoiesis networks [Citation32], and KLF15 binds to the promoter of KLF3 and promotes its expression in bovine adipocytes [Citation33]. Therefore, this study examined the expression changes of several members of KLFs after silencing the KLF9 gene in intramuscular and subcutaneous preadipocytes in goats. The expression of KLF2 and KLF5 was significantly up-regulated in both subcutaneous and intramuscular preadipocytes of goats. However, there are also interesting points, such as differentially up-regulating the expression of KLF3 and KLF7 and down-regulating KLF17 in intramuscular preadipocytes, up-regulating KLF1 and KLF17 and down-regulating the expression of KLF4, KLF12 and KLF13 genes in subcutaneous adipocytes. Therefore, it can be suggested that KLF9 may have a cascade-regulated relationship with these KLFs. Interestingly, it was found that many negative regulatory transcription factors were up-regulated after silencing the KLF9 gene, such as KLF2 [Citation34], KLF3 [Citation35], and KLF7 [Citation36]. It is speculated that it may be due to the following reasons: (1) Adipocyte differentiation was regulated by a large number of genes through transcriptional cascades pattern, while KLFs are only part of the many transcriptional regulators. (2) The same gene plays different roles in different type cells. For example, the gene promotes the differentiation of 3T3-L1 adipocytes by inducing the C/EBPβ gene [Citation37], but endogenous KLF4 has no effect on its differentiation [Citation38]. However, in the goat muscle, KLF4 plays an inhibitory role in the differentiation of somatic adipocytes (data not published). (3) Members of KLFs are involved in adipocyte differentiation, such as PPARγ and C/EBPs, by regulating differentiation marker genes [Citation32]. Therefore, this study determined that the cascade regulation of KLFs is involved during adipocyte differentiation including KLF9, and provided basic data for further elucidating the role of KLF9 in adipocyte differentiation in different parts of goat body.
In summary, our data indicates that KLF9 is a key transcription factor in regulating the differentiation of both intramuscular and subcutaneous adipocytes in goats, but possibly through differential mechanisms. KLF9 may be involved in subcutaneous adipocyte differentiation by regulating C/EBPβ and LPL genes, and may participate in intramuscular adipocyte differentiation by regulating aP2, C/EBPα, and Pref-1 genes. The results provide an important basis for further revealing the regulatory mechanisms on the differentiation of subcutaneous and intramuscular adipocytes in goat.
Author contribution
Yaqiu Lin and Yong Wang conceived and designed the experiments; Qing Xu performed the experiments, analyzed and interpreted the data; J.Z. assisted data analysis and interpretation; and Qing Xu wrote the manuscript; Wenlin Bai proofread this article and modified the statement critically. All authors approved the final version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Henry SL, Bensley JG, Wood-Bradley RJ, et al. White adipocytes: more than just fat depots. Int J Biochem Cell Biol. 2012;44:435–440.
- Lowe CE, O’rahilly S, Rochford JJ. Adipogenesis at a glance. J Cell Sci. 2011;124(Pt 16):2681–2686.
- White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318(1–2):10–14.
- Hocquette JF, Gondret F, Baeza E, et al. Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal. 2010;4(2):303–319.
- Zhang GH, Lu JX, Chen Y, et al. Comparison of the adipogenesis in intramuscular and subcutaneous adipocytes from Bamei and Landrace pigs. Biochem Cell Biol. 2014;92(4):259–267.
- Zhang HX, Zhu XT, Shu G, et al. Differential mRNA expression profiles of porcine intramuscular preadipocytes compared with subcutaneous preadipocytes during differentiation. Sci Agric Sin. 2008;11:3760–3768. in Chinese.
- Huang WL, Zhang XX, Li Y, et al. Identification of differentially expressed genes between subcutaneous and intramuscular adipose tissue of large white pig using RNA-seq. Hereditas. 2017;39(6):501–511. in Chinese.
- Bong JJ, Cho KK, Baik M. Comparison of gene expression profiling between bovine subcutaneous and intramuscular adipose tissues by serial analysis of gene expression. Cell Biol Int. 2009;34(1):125–133.
- Zhou G, Wang S, Wang Z, et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Sci. 2010;86(2):440–450.
- Pearson R, Jackb B, Lee SHY, et al. Co-regulator interactions in Krüppel-like factor transcriptional programs. Bio Krüppel-like Factors. 2009;4:51–64.
- Mcconnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381.
- Kimura H, Fujimori K. Activation of early phase of adipogenesis through Krüppel-like factor KLF9-mediated, enhanced expression of CCAAT/enhancer-binding protein β in 3T3-L1 cells. Gene. 2014;534(2):169–176.
- Pei H, Yao Y, Yang Y, et al. Kruppel-like factor KLF9 regulates PPARgamma transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011;18(2):315–327.
- Escalona-Nandez I, Guerrero-Escalera D, Estanes-Hernandez A, et al. The activation of peroxisome proliferator-activated receptor gamma is regulated by Kruppel-like transcription factors 6 & 9 under steatotic conditions. Biochem Biophys Res Commun. 2015;458(4):751–756.
- Peng Y, Chen FF, Ge J, et al. miR-429 inhibits differentiation and promotes proliferation in porcine preadipocytes. Int J Mol Sci. 2016;17(12):E2047.
- Jiang S, Wei H, Song T, et al. KLF13 promotes porcine adipocyte differentiation through PPARγ activation. Cell Biosci. 2015;5:28.
- Qing X, Sen L, Yong W, et al. Fibroblast growth factor 10 (FGF10) promotes the adipogenesis of intramuscular preadipocytes in goat. Mol Biol Rep. 2018;45(6):1881–1888.
- Xiong Y, Xu Q, Lin S, et al. Knockdown of LXRα inhibits goat intramuscular preadipocyte differentiation. Int J Mol Sci. 2018;19(10):E3037.
- Cai R, Sun Y, Qimuge N, et al. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(4):420–432.
- Zhao Y, Lin YQ, Zhu JJ, et al. Cloning and tissue expression profile of KLF9 gene in goat. Acta Agric Boreali-Sinica. 2017;32(6):106–112. in Chinese.
- Eaton SA, Funnell AP, Sue N, et al. A network of Kruppel-like factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283(40):26937–26947.
- Funnell AP, Maloney CA, Thompson LJ, et al. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol Cell Biol. 2007;27(7):2777–2790.
- Rosen E, Eguchi J, Xu Z. Transcriptional targets in adipocyte biology. Expert Opin Ther Targets. 2009;13(8):975–986.
- Farrmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273.
- White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318(12):10–14.
- Sun GR. Zhang M, Sun JW, et a. Krüppel-like factor KLF9 inhibits chicken intramuscular preadipocyte differentiation. Br Poult Sci. 2019;60(6):790–797.
- Mori T, Sakaue H, Iguchi H, et al. Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280(13):12867–12875.
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273.
- Siersbaek R, Nielsen R, Mandurp S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab. 2006;23(2):56–64.
- Wang Y, Kim KA, Kim JH, et al. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J Nutr. 2006;136(12):2953–2956.
- Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24(6):236–240.
- Wu Z, Wang S. Role of kruppel-like transcription factors in adipogenesis. Dev Biol. 2013;373(2):235–243.
- Guo H, Khan R, Raza SHA, et al. KLF15 promotes transcription of KLF3 gene in bovine adipocytes. Gene. 2018;659:77–83.
- Banerjee SS, Feinberg MW, Watanabe M, et al. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278(4):2581–2584.
- Sue N, Jack BH, Eaton A, et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveal s a role in adipogenesis. Mol Cell Biol. 2008;28(12):3967–3978.
- Kawamura Y, Tanaka Y, Kawamori R, et al. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006;20(4):844–856.
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7(4):339–347.
- Park YK, Wang L, Giampietro A, et al. Distinct roles of transcription factors KLF4, Krox20, and peroxisome proliferator-activated receptor γ in adipogenesis. Mol Cell Biol. 2017;37(2):e00554–16.