Abstract
Purpose
To investigate whether mutations in the minichromosome maintenance complex component (MCM) family genes were present in patients with polycystic ovary syndrome (PCOS) of Chinese descent.
Methods
A total of 365 Chinese patients with PCOS and 860 women without PCOS as control who underwent with assisted reproductive technology were enrolled. Genomic DNA was extracted from the peripheral blood of these patients for PCR and Sanger sequencing. The potential damage of these mutations/rare variants was analyzed through evolutionary conservation analysis and bioinformatic programs.
Results
Twenty-nine missense or nonsense mutations/rare variants in the MCM genes were identified in 365 patients with PCOS (7.9%, 29/365), all these mutations/rare variants were predicted to be ‘disease causing’ by SIFT and PolyPhen2 programs. Among those, four mutations were reported here for the first time, p.S7C (c.20C > G) in MCM2 (NM_004526.3), p.K350R (c.1049A > G) in MCM5 (NM_006739.3), p.K283N (c.849G > T) in MCM10 (NM_182751.2), and p.S1708F (c.5123C > T) in MCM3AP (NM_003906.4). All of these novel mutations were not found in our 860 control women, or also absent in public databases. In addition, the evolutionary conservation analysis results suggested that these novel mutations caused highly conserved amino acid substitutions among 10 vertebrate species.
Conclusion
This study identified a high frequency of potential pathogenic rare variants/mutations in MCM family genes in Chinese women with PCOS, which further expands the genotype spectrum in PCOS.
Introduction
Polycystic ovary syndrome (PCOS) is a common reproductive endocrine disorder of females, affecting at least 10% of reproductive-aged women [Citation1,Citation2]. It is typically characterized by the presence of at least two of the three cardinal features of clinical and/or biochemical hyperandrogenemia (high circulating androgen levels), ovulatory dysfunction, and polycystic ovaries [Citation3]. PCOS leads to diverse clinical implications including reproductive, metabolic, and psychological features [Citation4]. The etiology of PCOS has not been well understood. Generally, multiple studies show that both environmental and genetic factors play important roles in the etiology of PCOS [Citation5–7].
Familial clustering and twin studies indicate that PCOS is a complex genetic disease, due to its high inheritance rates and heterogeneous phenotypes [Citation8]. Multiple new risk loci and candidate genes have been identified for PCOS by Genome Wide Association Studies (GWAS) [Citation8,Citation9–13], but less than 10% of heritability have been explain through these findings. Therefore, we could speculate that different phenotypes and subphenotypes are caused by rare unique genetic variants.
The minichromosome maintenance (MCM) family is highly conserved in vertebrates and comprises MCM2 through MCM10. These proteins play essential roles in DNA replication and cell cycle progression. Indeed, MCM proteins not only interact with S-phase checkpoint regulators, but also with components of DNA repair pathway [Citation14–16]. The MCM 2-7 complex is instrumental for cell-cycle control, an operation that occurs during the G1 and G2 phases of mitosis [Citation14,Citation15]. MCM8 complexes with MCM9 and has a role in DNA repair and genome instability [Citation17–19]. MCM10 involves in DNA replication and chromosomal instability [Citation16,Citation20]. Recently, studies showed that dysfunctional mutations in MCM8 and MCM9 could lead to premature ovarian failure (POF) and primary ovarian insufficiency (POI) [Citation19,Citation21–23]. Considering the shared biological features between POF/POI and PCOS, such as dysregulation of steroid hormones, and ovarian dysfunction, moreover, some mutations in POF/POI also exist in PCOS, such as AMH, FSHR, GDF9, and BMP15 [Citation24–26], thus, we speculate that mutations/rare variants of MCMs might exist in Chinese patients with PCOS.
Here, we performed Sanger sequencing of MCMs in 365 Chinese women with PCOS and identified twenty-nine missense mutations (7.9%, 29/365), of which, four are novel mutations. None of these novel mutations was found in public databases or in our 860 control women. In addition, the evolutionary conservation analysis results suggested that these novel mutations caused highly conserved amino acid substitutions among vertebrate species. In brief, we identified novel mutations in MCM family genes that are potentially causative for Chinese women with PCOS, which further expands the genotype spectrum in PCOS.
Materials and methods
Study population
A total of 365 patients with PCOS and 860 control women were recruited from Jiangxi Provincial Maternal and Child Health Hospital (Nanchang, China) between October 2016 and September 2018. The clinical cases of PCOS were confirmed by the guidelines of the 2003 Rotterdam Criteria [Citation27]. Women with oligimenorrhea or hyperandrogenism of other causes, including congenital adrenal hyperplasia, 21-hydroxylase-deficient non-classic adrenal hyperplasia, hyperprolactinemia, Cushing’s syndrome, androgen-secreting tumors, or androgenic/anabolic drug use or abuse, were excluded from this study. Women without PCOS, including male infertility or tubal factor infertility, were recruited as the control group for the same period. All the controls had regular menstrual cycles, normal androgen levels, and had no history of cancer, diabetes, galactorrhea, or any endocrine or systemic disease that could affect reproductive physiology. The available clinical data of the patients is presented in . In addition, a consent form for the study was provided by each participant, and ethical approval for the study was granted by the Institutional Research Ethics Committee of Jiangxi Provincial Maternal and Child Healthcare Hospital. Peripheral blood was taken from the participants for DNA extraction in an anticoagulant-treated tube.
Table 1. Clinical data of the patients.
DNA extraction
Total DNA extraction from each participant was performed with a DNeasy Blood & Tissue Kit (Qiagen, cat. no. 69504), following the manufacturer’s instructions. The extracted DNA was dissolved in 100 μL of TE buffer. The DNA was quantified by NanoDrop (Thermo Scientific, Waltham, USA), and DNA quality was checked by determining optical density (OD) at 260 and 280 nm. Then the DNA samples were stored at –80 °C until use.
PCR and Sanger sequencing
According to the entire coding regions and the adjacent exon/intron boundaries of each MCM family (MCM2-10, MCMDC2, and MCM3AP) member, primers were designed using Primer3 online tool. The primers list is presented in Supplemental Table 1.
The PCR was carried out in a final reaction volume of 25 μL containing ∼50 ng total DNA, 12 μL of Ampliqon master mix (TaKaRa, cat. no: RR320), 10 pmol of forward primer and 10 pmol of reverse primer. The PCR program involved an initial pre-denaturation step at 95 °C for 3 min, followed by 30 cycles, including a denaturation step at 95 °C for 30 s, a primer annealing step at different temperatures (52–60 °C, Supplemental Table 1) for 30 s, and an extension step at 72 °C for 30 s, final extension step at 72 °C for 7 min. Thermal Cycler 2720 (Applied Biosystems, Thermo Fisher Scientific, Inc.) was used for performing PCR. Briefly, 2 μL of PCR products were resolved by 2% agarose gel for evaluating the PCR products length. The PCR products were then purified and sequenced on an ABI Prism 3730 DNA sequencer (Applied Biosystems, Thermo Fisher Scientific, Inc.). The sequencing chromatograms were analyzed with DNAStar software. All the variants were confirmed by three independent PCR runs and sequenced in both forward and reverse strands. SIFT (http://sift.jcvi.org/) and PolyPhen2 (http://genetics.bwh.harvard.edu/), the online bioinformatic programs, were used to analyze the disease-causing potential of the identified variants.
Evolutionary conservation analysis
Evolutionary Conservation Analysis was conducted on the ClustalW2 web site (www.clustal.org/clustal2) among 10 vertebrate species from Ensembl database, to predict the potential pathogenicity of the novel variants.
Protein structural modeling
The available three-dimensional (3D) structures of human MCM5 and MCM10 protein were obtained from the SWISS-MODEL (http://swissmodel.expasy.org/). Then, the changes of 3D structure between the wild type and mutant proteins were compared by DeepView Swiss-PdbViewer 4.0 software, respectively.
Statistical analysis
SPSS 18.0 software (IBM) was used for data analysis. The continuous data were described as the mean ± standard deviation (SD). Categorical data was tested by Pearson’s χ2 test or Fisher’s exact test. All the P values were two-sided, and p < .05 was considered to indicate a statistical difference.
Results
Four novel mutations of MCM family genes identified in PCOS samples
Through Sanger sequencing in 365 patients with PCOS, four novel missense variations and twenty-five known single nucleotide polymorphisms (SNPs) in MCM family genes were identified (7.95%, 29/365), while thirty-four out of 860 control women were identified who carried those SNPs (3.95%, 34/860) (p = .0109) ( and ). Four novel missense variations, p.S7C (c.20C > G) in MCM2 (NM_004526.3), p.K350R (c.1049A > G) in MCM5 (NM_006739.3), p.K283N (c.849G > T) in MCM10 (NM_182751.2), and p.S1708F (c.5123C > T) in MCM3AP (NM_003906.4), were absent both in the ExAC (http://exac.broadinstitute.org/) database or 860 control women without PCOS (). In addition, all of the mutations were found to be harmful to the function of its proteins according to the prediction of SIFT and PolyPhen2 software programs (). The domain architecture of MCM proteins and the position of the mutations found in our study are shown in .
Figure 1. The sequence chromatograms of MCM family genes variations identified in Chinese patients with PCOS. The arrow refers to the location of the mutation, compared with normal control sequences.
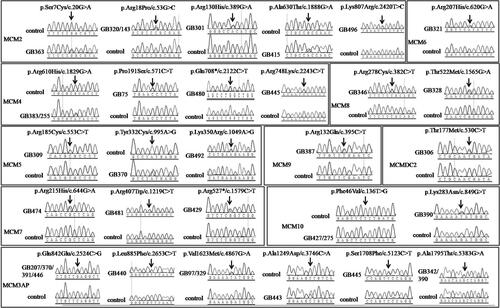
Figure 2. The schematic diagram of the MCM family genes. The arrow refers to the location of the mutations identified in patients with PCOS. The red arrows show the novel mutations identified in our study.
Table 2. Genetic variants of MCM gene family.
Evolutionary conservation analysis and protein structural modeling
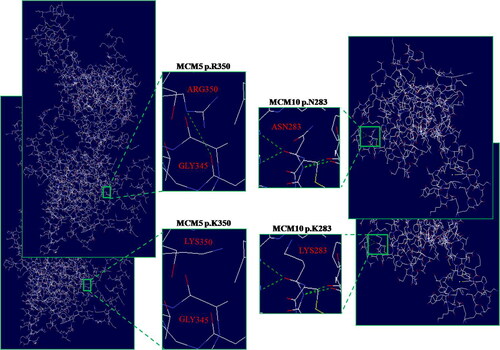
We performed evolutionary conservation analysis of the four novel mutations in the MCM family genes. The results suggested that these mutations were highly conserved among 10 vertebrate species, including monkeys, horses, pandas, cats, sheep, pigs, and so on (). Furthermore, we analyzed the MCM5 and MCM10 3D structure changes between the wild type and mutant proteins respectively, duo to only the 3D structure of MCM5 and MCM10 could be obtained from the SWISS-MODEL, while others could not. The results of protein structural modeling showed that both the MCM5 p.K350R and MCM10 p.K283N mutations caused the structure changes of MCM5 and MCM10 protein, respectively ().
Figure 3. The evolutionary conservation analysis of the four novel mutations in the MCM family genes. These results suggested that these mutations were highly conserved among 10 vertebrate species from Human to Pig.

Figure 4. Structural difference between wild type and mutant type proteins. The proteins structural of wild type and mutany type were modeled based on the crystal model of human MCM5 and MCM10 proteins, respectively. The enlarged image shows that both the MCM5 p.K350R and MCM10 p.K283N mutations caused the structure changes of MCM5 and MCM10 protein, respectively.
Discussion
MCMs were first identified in Saccharomyces cerevisiae as imperative factors in the maintenance of extrachromosomal DNA [Citation28]. Prior studies had shown that MCMs were functionally related to double-strand DNA unwinding, DNA replication control, and DNA damage repair [Citation29]. Genomic instability resulting from MCMs variations has been elucidated to have an association with multiple diseases, including POI and POF, which are the major causes of infertility [Citation19,Citation21–23]. Based on the multiple mutations in MCM family genes identified in infertile patients [Citation22,Citation30–32], we speculated that women with PCOS might also harbor mutations in MCM family genes.
PCOS is a complex disorder, which is the leading cause of anovulatory infertility. The pathogenesis of PCOS is still not fully understood. Epidemiological studies have shown that PCOS is closely associated with genetic factors [Citation8]. Searching for major genes of PCOS is the key to clarifying its molecular pathogenesis. In a recent study with a three-generation family revealed that variants of SCARB1 and INSR were found to be pathogenic to Indian PCOS women [Citation33]. Moreover, in the study by Feng et al. on 150 people with PCOS and 300 controls indicated MTHFR A1298C and MTRR A66G were associated with PCOS, and MTHFR A1298C might affect the risk of PCOS by influencing the homocysteine level [Citation34]. In our present study, twenty-nine missense mutations in MCM family genes were identified in 365 of Han Chinese patients with PCOS, among which, P.S7C (c.20C > G) in MCM2 (NM_004526.3), p.K350R (c.1049A > G) in MCM5 (NM_006739.3), p.K283N (c.849G > T) in MCM10 (NM_182751.2), and p.S1708F (c.5123C > T) in MCM3AP (NM_003906.4) were reported here for the first time.
In eukaryotic cells, the MCM family plays a crucial role in DNA replication. MCM2 to MCM7 (MCM2–7) form a heterohexameric complex, which functions as replicative DNA helicase to unwind the DNA duplex template during DNA replication [Citation25]. Previous studies have shown that MCM 10 is one of the replication initiation factors and binds to MCM2-7 to activate their helicase activity [Citation20,Citation35,Citation36], and MCM3AP can bind to the MCM3 protein involved in initiation of DNA replication [Citation37,Citation38]. In addition, MCM8 and MCM9 are not only involved in DNA replication, but also participate in homologous recombination (HR) during meiosis and DNA double-stranded break (DSB) repair, which are crucial for gonadal development and ovarian function [Citation17,Citation18]. Failure or incompletion of DNA replication causes genome instability, potentially resulting in cell death, cancer development, or genetic disorder. Mutations/rare variants in MCM genes have been found to be associated with various human diseases, such as MCM2 (R44C) mutation can lead to non-syndromic sensorineural hearing loss [Citation39], MCM4 p.G364R mutation was detected in human skin cancer and p.G486D mutation was detected in endometrial carcinoma [Citation40,Citation41], MCM5 (T466I) mutation can lead to Meier–Gorlin syndrome-8 (MGORS8) [Citation42], p.A265T mutation in MCM7 was detected in primary microcephaly [Citation43], and mutations in MCM8 or MCM9 were found with functional impairment in DNA damage repair in POI [Citation32,Citation44,Citation45]. Significantly, most of the mutations/rare variants in MCM genes that we identified were predicted to be “disease causing” by SIFT and PolyPhen2 programs. Meanwhile, the primary structures of human MCM proteins showed the locations of the mutations, and most were located in critical domains, which might interfere with physiological function and activity of proteins [Citation28,Citation34,Citation46]. These results suggested that these mutations might be functionally important.
Taken together, we speculated that most of these mutations of MCM family genes might participate positively in the progression of PCOS. It has been shown that MCM genes may activate the ERK pathway. It is thought that the ERK-1 and ERK-2 pathways may be related to the pathogenesis of PCOS [Citation47,Citation48]. Therefore, MCM genes may also contribute to the PCOS pathogenesis through this pathway. However, it should be noted that this observation should be treated cautiously as the sample size was too small, and the causal relationship between these potential candidate sites and PCOS needs to be verified by further functional experiments.
In summary, we identified high frequency of potential pathogenic rare variants/mutations in MCM family genes in Chinese women with PCOS, which further expands the genotype spectrum in PCOS.
Supplemental Material
Download MS Word (36.9 KB)Acknowledgment
We sincerely thank all the patients for donating their peripheral blood.
Disclosure statement
The authors report there are no competing interests to declare.
Availability of data and materials
Not applicable.
Additional information
Funding
References
- Azziz R, Carmina E, Chen Z, et al. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:1.
- Ajmal N, Khan SZ, Shaikh R. Polycystic ovary syndrome (PCOS) and genetic predisposition: a review article. Eur J Obstet Gynecol Reprod Biol X. 2019;3:100060.
- Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2004;19(1):41–8.
- Teede H, Deeks A, Moran L. Polycystic ovary syndrome: a complex condition with psychological, reproductive and metabolic manifestations that impacts on health across the lifespan. BMC Med. 2010;8:41.
- Merkin SS, Phy JL, Sites CK, et al. Environmental determinants of polycystic ovary syndrome. Fertil Steril. 2016;106(1):16–24.
- Dadachanji R, Shaikh N, Mukherjee S. Genetic variants associated with hyperandrogenemia in PCOS pathophysiology. Genet Res Int. 2018;2018:7624932.
- Dapas M, Sisk R, Legro RS, et al. Family-based quantitative trait meta-analysis implicates rare noncoding variants in DENND1A in polycystic ovary syndrome. J Clin Endocrinol Metab. 2019;104(9):3835–3850.
- Day F, Karaderi T, Jones MR, et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14(12):e1007813.
- Hayes MG, Urbanek M, Ehrmann DA, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in european ancestry populations. Nat Commun. 2015;6:7502.
- Day FR, Hinds DA, Tung JY, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464.
- Shi Y, Zhao H, Shi Y, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44(9):1020–1025.
- Chen ZJ, Zhao H, He L, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43(1):55–59.
- Verdiesen RMG, Van der Schouw YT, Van Gils CH, et al. Genome-wide association study meta-analysis identifies three novel loci for circulating anti-Müllerian hormone levels in women. Hum Reprod. 2022;37(5):1069–1082.
- Masai H, You Z, Arai K. Control of DNA replication: regulation and activation of eukaryotic replicative helicase, MCM. IUBMB Life. 2005;57(4–5):323–335.
- Das SP, Rhind N. How and why multiple MCMs are loaded at origins of DNA replication. Bioessays. 2016;38(7):613–617.
- Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell. 2007;18(10):4085–4095.
- Nishimura K, Ishiai M, Horikawa K, et al. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell. 2012;47(4):511–522.
- Lutzmann M, Grey C, Traver S, et al. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 2012;47(4):523–534.
- AlAsiri S, Basit S, Wood-Trageser MA, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest. 2015;125(1):258–262.
- Looke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31(3):291–305.
- Vetro A, Savasta S, Russo Raucci A, et al. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur J Hum Genet. 2017;25(5):646–650.
- Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, et al. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52(6):391–399.
- Fauchereau F, Shalev S, Chervinsky E, et al. A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency. Clin Genet. 2016;89(5):603–607.
- Qin C, Yuan Z, Yao J, et al. AMH and AMHR2 genetic variants in chinese women with primary ovarian insufficiency and normal age at natural menopause. Reprod Biomed Online. 2014;29(3):311–318.
- Kumar R, Alwani M, Kosta S, et al. BMP15 and GDF9 gene mutations in premature ovarian failure. J Reprod Infertil. 2017;18(1):185–189.
- Sproul K, Jones MR, Mathur R, et al. Association study of four key folliculogenesis genes in polycystic ovary syndrome. BJOG. 2010;117(6):756–760.
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81(1):19–25.
- Ishimi Y. Regulation of MCM2-7 function. Genes Genet Syst. 2018;93(4):125–133.
- Yu S, Wang G, Shi Y, et al. MCMs in cancer: prognostic potential and mechanisms. Anal Cell Pathol (Amst). 2020;2020:3750294.
- Alvarez-Mora MI, Todeschini AL, Caburet S, et al. An exome-wide exploration of cases of primary ovarian insufficiency uncovers novel sequence variants and candidate genes. Clin Genet. 2020;98(3):293–298.
- Chen S, Wang G, Zheng X, et al. Whole-exome sequencing of a large chinese azoospermia and severe oligospermia cohort identifies novel infertility causative variants and genes. Hum Mol Genet. 2020;29(14):2451–2459.
- Zhang YX, He WB, Xiao WJ, et al. Novel loss-of-function mutation in MCM8 causes premature ovarian insufficiency. Mol Genet Genomic Med. 2020;8(4):e1165.
- Janani DM, Ramasubramanyan S, Chellappa V, et al. Whole exome and targeted sequencing reveal novel mutations associated with inherited PCOS condition in an Indian cohort. J Hum Genet. 2023;68(1):39–46.
- Feng W, Zhang Y, Pan Y, et al. Association of three missense mutations in the homocysteine-related MTHFR and MTRR gene with risk of polycystic ovary syndrome in Southern chinese women. Reprod Biol Endocrinol. 2021; 719(1):5.
- Deegan TD, Yeeles JT, Diffley JF. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016;35(9):961–973.
- Izumi M, Mizuno T, Yanagi KI, et al. The Mcm2-7-interacting domain of human mini-chromosome maintenance 10 (Mcm10) protein is important for stable chromatin association and origin firing. J Biol Chem. 2017;292(31):13008–13021.
- Abe E, Kuwahara K, Yoshida M, et al. Structure, expression, and chromosomal localization of the human gene encoding a germinal center-associated nuclear protein (GANP) that associates with MCM3 involved in the initiation of DNA replication. Gene. 2000;255(2):219–227.
- Gao J, Wang Q, Dong C, et al. Whole exome sequencing identified MCM2 as a novel causative gene for autosomal dominant nonsyndromic deafness in a chinese family. PLoS One. 2015;10(7):e0133522.
- Takei Y, Assenberg M, Tsujimoto G, et al. The MCM3 acetylase MCM3AP inhibits initiation, but not elongation, of DNA replication via interaction with MCM3. J Biol Chem. 2002;277(45):43121–43125.
- Shimi Y, Irie D. G364R mutation of MCM4 detected in human skin cancer cells affects DNA helicase activity of MCM4/6/7 complex. J Biochem. 2015;157(6):561–569.
- Tatsumi R, Ishimi Y. An MCM4 mutation detected in cancer cells affects MCM4/6/7 complex formation. J Biochem. 2017;161(3):259–268.
- Wood-Trageser MA, Gurbuz F, Yatsenko SA, et al. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95(6):754–762.
- Ravindran E, Gutierrez de Velazco C, Ghazanfar A, et al. Homozygous mutation in MCM7 causes autosomal recessive primary microcephaly and intellectual disability. J Med Genet. 2022;59(5):453–461.
- Guo T, Zheng Y, Li G, et al. Novel pathogenic mutations in minichromosome maintenance complex component 9 (MCM9) responsible for premature ovarian insufficiency. Fertil Steril. 2020;113(4):845–852.
- Wang F, Guo S, Li P. Two novel mutations in the MCM8 gene shared by two chinese siblings with primary ovarian insufficiency and short stature. Mol Genet Genomic Med. 2020;8(9):e1396.
- Sedghi M, Moslemi AR, Cabrera-Serrano M, et al. Recessive Charcot-Marie-Tooth and multiple sclerosis associated with a variant in MCM3AP. Brain Commun. 2019;1(1):fcz011.
- Wang Y, Chen H, Zhang J, et al. MCM family in gastrointestinal cancer and other malignancies: from functional characterization to clinical implication. Biochim Biophys Acta Rev Cancer. 2020;1874(2):188415.
- Guney G, Taşkın MI, Sener N, et al. The role of ERK-1 and ERK-2 gene polymorphisms in PCOS pathogenesis. Reprod Biol Endocrinol. 2022;20(1):95.
