?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Atom Search Optimisation (ASO) is a recently proposed metaheuristic algorithm that has proved to work effectively on several benchmark tests. In this paper, we propose the binary variants of atom search optimisation (BASO) for wrapper feature selection. In the proposed scheme, eight transfer functions from S-shaped and V-shaped families are used to convert the continuous ASO into the binary version. The proposed BASO approaches are employed to select a subset of significant features for efficient classification. Twenty-two well-known benchmark datasets acquired from the UCI machine learning repository are used for performance validation. In the experiment, the BASO with an optimal transfer function that contributes to the best classification performance is presented. The particle swarm optimisation (PSO), binary differential evolution (BDE), binary bat algorithm (BBA), binary flower pollination algorithm (BFPA), and binary salp swarm algorithm (BSSA) are used to evaluate the efficacy and efficiency of proposed approaches in feature selection. Our experimental results reveal the superiority of proposed BASO not only in high prediction accuracy but also in the minimal number of selected features.
1. Introduction
With the rapid growth of information technology, the amount of data is evolving exponentially. Ideally, all the information offered by the features is meaningful. Many pattern recognition applications, extracting the features to describe the target concept in the classification tasks. However, the dataset normally contains irrelevant and redundant features, which significantly degrades the performance of the classification model (Wang et al., Citation2018; Peng et al., Citation2005). The excessive number of features not only introduces the extra computational complexity but also increases the prediction error (Labani et al., Citation2018).
Feature selection is one of the effective ways to resolve the issues above. Briefly, feature selection attempts to select a small subset of significant features that can maintain or improve the prediction accuracy (Liu et al., Citation2018). In general, feature selection can be categorised into wrapper and filter approaches. Wrapper approaches utilise a learning algorithm to evaluate the optimal feature subset (Mafarja & Mirjalili, Citation2017). On the one hand, filter approaches use the information theory and statistical analysis over feature space to remove the redundant and irrelevant features (Labani et al., Citation2018). In comparison with wrapper, filter approaches can usually work faster, and they are independent of the learning algorithm. However, wrapper approaches can often achieve better classification results (Xue et al., Citation2014). Thus, this study focuses on the wrapper feature selection.
The main goal of feature selection is to improve classification performance and reduce the number of features. Thus it can be considered as a combinatorial optimisation task (Faris et al., Citation2018). Wrapper feature selection is performed using the metaheuristic algorithms such as genetic algorithm (GA) (Huang & Wang, Citation2006), ant colony optimisation (ACO) (Al-Ani, Citation2005), and binary particle swarm optimisation (BPSO) (Chuang et al., Citation2008), binary grey wolf optimisation (BGWO) (Emary et al., Citation2016a), binary salp swarm algorithm (BSSA) (Faris et al., Citation2018), binary tree growth algorithm (BTGA) (Too et al., Citation2018), multi-verse optimiser (BMVO) (Faris et al., Citation2017), and binary differential evolution (BDE) (Zorarpacı & Özel, Citation2016). Previous works showed that metaheuristic algorithms were having high potential when solving the feature selection problem, which made them received a lot of attention from researchers. However, according to the No Free Lunch theorem (NFL), there is no universal metaheuristic algorithm that can solve all the feature selection problems effectively (Wolpert & Macready, Citation1997). Therefore, more and more new algorithms are required for efficient feature selection.
In the past study, Wang et al. (Citation2017) proposed a modified binary-coded ant colony optimisation (MBACO) in which the GA was used to generate a population of high-quality initial solutions for image classification. Rodrigues et al. (Citation2014) applied the binary bat algorithm (BBA) with the optimum-path forest as part of evaluations for feature selection. Moreover, Mafarja et al. (Citation2019) introduced the binary grasshopper optimisation algorithm (BGOA) for feature selection tasks. The authors indicated that the utilisation of S-shaped and V-shaped transfer functions allowed the BGOA to search around the binary search space. Another study shows the implementation of transfer function into the binary butterfly optimisation algorithm (BBOA) can effectively tackle the feature selection problems (Arora & Anand, Citation2019). More recent studies of wrapper feature selection can be found in (Al-Tashi et al., Citation2019; Emary et al., Citation2016b; Faris et al., Citation2018; Mafarja et al., Citation2018; Mirhosseini & Nezamabadi-pour, Citation2018; Pashaei & Aydin, Citation2017).
Atom Search Optimisation (ASO) is a recently established metaheuristic algorithm inspired by the concept of molecular dynamics (Zhao et al., Citation2019a). To date, ASO has attracted the attention of various researchers due to its efficacy in solving global optimisation for different applications. In comparison with the Water Drop Algorithm (WDA) (Siddique & Adeli, Citation2014), Particle Swarm Optimisation (PSO) (Kennedy, Citation2011), GA and Gravitational Search Algorithm (GSA) (Rashedi et al., Citation2009), ASO can usually find promising solutions. Hence, ASO can be a potential metaheuristic algorithm for other real-world applications such as feature selection. To the best of our knowledge, there is no study to apply the ASO for feature selection, which becomes the motivation of this work. This encourages us to develop the binary version of ASO to tackle the feature selection problem in classification tasks.
In this study, we propose the new binary version of atom search optimisation (BASO) for wrapper feature selection. The BASO integrates the S-shaped or V-shaped transfer function, which allows the search agent to move on the binary search space. Twenty-two benchmark datasets acquired from the UCI machine learning repository are used to validate the performance of proposed BASO, and the results are compared with the other five recent and popular algorithms. From the experiment, it shows that BASO is highly capably in evaluating the optimal feature subset, which leads to promising results.
The rest of paper is organised as follows: Section 2 details the standard atom search optimisation. Section 3 describes the proposed binary atom search optimisation approaches. Section 4 depicts the application of proposed approaches for feature selection. Section 5 discusses the findings of the experiments. Finally, Section 6 concluded the findings of the research work.
2. The atom search optimisation
Atom Search Optimisation (ASO) is a new metaheuristic algorithm proposed by Zhao and his colleagues in 2019 (Zhao et al., Citation2019a). The ASO mimics the basic concept of molecular dynamics and movement principle of atoms such as characteristics of the potential function, interaction force, and geometric constraint force. In ASO, the population of solutions is called atoms, and each atom maintains two vectors, namely, position and velocity.
All the atoms are moving at all times by following the movement principle. Mathematically, the acceleration of atom is defined as:
(1)
(1) where F is the interaction force, G is the constraint force, and m is the mass of the atom.
The interaction force between the ith atom and jth atom is described by the Lennard-Jones (L-J) potential as:
(2)
(2) and
(3)
(3) where ε is the depth of potential, σ is the length scale, r is the distance between two atoms, d is the dimension and t is the current iteration. However, Equation (3) cannot directly apply for optimisation tasks. Thus, a revised version of Equation (3) is designed as follow:
(4)
(4) where η is the depth function to regulate the attraction or repulsion region, and it can be expressed as:
(5)
(5) where α is the depth weight and T is the maximum number of iterations. The function h can be computed as follow:
(6)
(6) where hmax and hmin are the upper and lower limit of h, and they are set to 1.1 and 2.4, respectively (Zhao et al., Citation2019b). The length scale σt can be calculated as:
(7)
(7) and
(8)
(8) where K is the subset of atoms with better fitness values, g0 and µ are equal to 1.1 and 2.4, respectively. The g is the drift factor that controls the balance between exploration and exploitation, and it is defined as:
(9)
(9)
The total interaction force can be expressed as:
(10)
(10) where r is a random number in [0,1]. According to Newton’s third law, the force on the jth atom for the same pairwise interaction is the opposite of force on the ith atom as (Zhao et al., Citation2019b):
(11)
(11)
Furthermore, geometric constraint of the ith atom and constraint force can be expressed as follows:
(12)
(12)
(13)
(13) where Xbest is the position of the best atom, bi,bestis the fixed bond length between ith atom and the best atom, and λ is the Lagrangian multiplier. By making 2λ = λ, the constraint force can be represented as:
(14)
(14)
The Lagrangian multiplier can be expressed as:
(15)
(15) where β is the multiplier weight. Finally, the acceleration of the atom can be written as:
(16)
(16) where m is the mass of the atom, which can be calculated by
(17)
(17)
(18)
(18) where Fit is the fitness value. Considering the minimisation problem, the Fitbest and Fitworst are represented as:
(19)
(19)
(20)
(20) Then, the velocity and position of the atom are updated as follows:
(21)
(21)
(22)
(22) where Xi and Vi are the position and velocity of the ith atom, a is the acceleration, d is the dimension of search space, r1 is a random number in [0,1], and t is the current iteration.
In ASO, the number of best atoms in subset K is used to control the exploration and exploitation phase.
(23)
(23) where N is the number of atoms in the population. Initially, higher k promotes exploration. At the end of the iteration, lower k ensures higher exploitation. The pseudocode of ASO is demonstrated in Algorithm 1.
3. Binary version of atom search optimisation
In the continuous version of ASO, the atoms are moving around the search space with the continuous real domain. As for binary optimisation, the atoms are dealing with only two numbers (“1” or “0”). Correspondingly, a way should be found to use the velocity of the atom to change the position from “0” to “1” or vice versa. Among the early works, the transfer function has successfully utilised in binary versions of particle swarm optimisation, gravitational search algorithm, bat algorithm, tree growth algorithm and antlion optimiser (Emary et al., Citation2016b; Kennedy & Eberhart, Citation1997; Mirjalili et al., Citation2014; Rashedi et al., Citation2010; Too et al., Citation2018). Previous works show that transfer function is one of the powerful tools to convert the continuous optimisation algorithm into the binary version (Mirjalili & Lewis, Citation2013).
In this paper, we propose eight different binary variants of ASO (BASO). The proposed BASO approaches are classified into two groups, namely S-shaped family and V-shaped family. The former applies an S-shaped transfer function that enables the search agent to move in the binary search space. The latter uses a V-shaped transfer function that allows the atoms to perform the search on the binary search space.
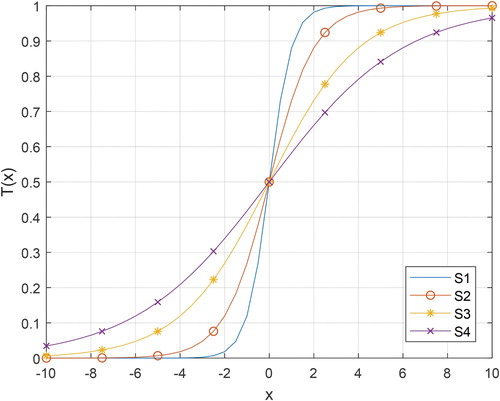
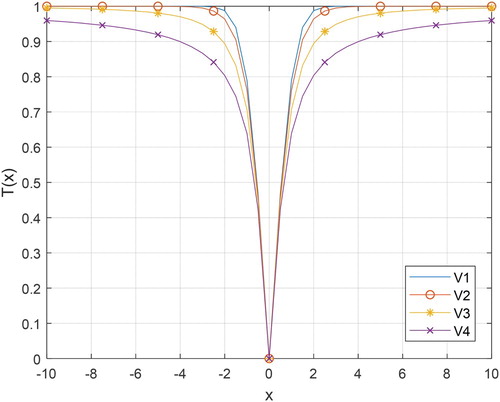
Table displays the S-shaped and V-shaped transfer functions with mathematical equations. In Table , the “erf” denotes the error function. The illustrations of the S-shaped and V-shaped families are shown in Figure and Figure , respectively. By integrating a transfer function into ASO, the binary version of ASO is achieved. In this framework, the velocity of the atom is first converted into probability using the S-shaped or V-shaped transfer function. Then, the position of the atom is updated.
Table 1. S-shaped and V-shaped transfer functions.
According to literature, the position of the search agent can be updated by using the Equation (Equation24(24)
(24) ) for S-shaped family or Equation (Equation25
(25)
(25) ) for V-shaped family, which converted the probability into binary representation (Kennedy & Eberhart, Citation1997; Mirjalili & Lewis, Citation2013; Rashedi et al., Citation2010).
(24)
(24)
(25)
(25) where Xi,d and Vi,d are the position and velocity of the ith atom at dth dimension, t is the current iteration, and rand is a random vector in [0,1]. As can be seen in Equations (Equation24
(24)
(24) ) and (Equation25
(25)
(25) ), the atom randomly changed its position based on the parameter rand. Taking Equation (Equation25
(25)
(25) ) as an example, if the rand is greater, then a higher value of T(V) is required for the atom to change its position. On the contrary, a lower value of T(V) enables the atom to maintain its position when the rand is greater. However, the atom can also change its position when the value of the rand is smaller. This indicates that the position of the atom is updated based on a fully random manner, which may lead to unsatisfactory performance. Thus, we propose the new updating rules as follow:
S-shape family
(26)
(26) V-shape family
(27)
(27)
The K1 and K2 are calculated as follows:
(28)
(28)
(29)
(29) where rand is a random vector in [0,1], Xi,d and Vi,d are the position and velocity of the ith atom at dth dimension, Xbest is the position of the best atom, and t is the current iteration. As can be seen in Equation (Equation26
(26)
(26) ), the parameter rand has removed and replaced with the K1 and K2. The atom no longer using a fully random manner to update its position. In our proposed scheme, if the value of T(V) is smaller (less than K1), then the atom set its new position to 0; else if the value of T(V) is medium (between K1 and K2), then the atom changes its position to 1; otherwise, the atom follows the position of the best atom for the next iteration. We involve the best atom in the proposed scheme to guide the atom move toward the global optimal.
The pseudocode of binary atom search optimisation (BASO) is illustrated in Algorithm 2. Firstly, the position and velocity of the N atoms are randomly initialised. Secondly, the fitness values of atoms are calculated, and the best atom is defined. In each iteration, for each atom, the mass is computed using Equations (17) and (18). Then, the interaction and constraint forces are determined using Equations (10) and (14), respectively. Next, the acceleration of the atom is computed as shown in Equation (16). The velocity of the atom is then updated using Equation (21). After that, the velocity of the atom is converted into probability value using the S-shaped or V-shaped transfer function. Later, the position of the atom is updated using Equation (26) or Equation (27). At the end of each iteration, the best atom is updated. The procedure is repeated iteratively until the maximum number of iterations is satisfied. Finally, the global best solution is achieved. The main differences between ASO and BASO are that there is an additional conversion step after velocity update and the position updating step.
4. Proposed binary atom search optimisation for feature selection
The proposed binary atom search optimizations are applied to solve the feature selection problem for classification tasks. Feature selection is a pre-processing step to evaluate the subset of significant features that can significantly enhance the prediction accuracy (Xue et al., Citation2014). From the optimisation perspective, the feature selection problem is considered as a binary combinatorial optimisation, which represents the solution in binary form. In feature selection, “1” means the feature is selected while “0” represents the unselected feature. Given a solution X = {1, 0, 1, 0, 0, 0, 1, 1, 1, 0}, it shows that total five features (1st, 3rd, 7th, 8th, and 9th features) are selected.
The optimal feature subset is determined based on two criteria: High classification accuracy and minimal feature size. In this study, the fitness function that used to evaluate the individual search agent is expressed as:
(30)
(30) and
(31)
(31) where ER is the error rate, |C| is the number of selected features, |F| is the total number of features in each dataset, and δ is used to control the influence of classification performance and feature size. Since the classification performance is considered to be the most important metric, we set δ to 0.99 (Emary et al., Citation2016a; Faris et al., Citation2018).
5. Experimental results and discussions
5.1. Data Description
In the experiment, twenty-two datasets acquired from the UCI repository are used to validate the performance of proposed approaches (UCI Machine Learning Repository). Table listed the utilised datasets. In this study, the K-fold cross-validation manner is applied for the performance evaluation. The data is divided into K equal parts, in which the K-1 parts are used for the training set, and the remaining part is used for the testing set. The procedure is repeated with K time by using different parts as the testing set. As for wrapper feature selection, a learning algorithm (classifier) is required to compute the error rate. According to (Emary et al., Citation2016b; Xue et al., Citation2014), k-nearest neighbour (KNN) with Euclidean distance and k = 5 is applied in this work. The KNN is chosen since it can usually achieve satisfactory performance with fast processing speed. In comparison with other classifiers, KNN not only simpler and faster but also easy to implement.
Table 2. List of used datasets.
5.2. Comparison algorithms and evaluation metrics
Five recent and popular feature selection methods, namely binary bat algorithm (BBA) (Mirjalili et al., Citation2014), particle swarm optimisation (PSO) (Mafarja et al., Citation2019), binary differential evolution (BDE) (Zorarpacı & Özel, Citation2016), binary salp swarm algorithm (BSSA) (Faris et al., Citation2018) and binary flower pollination algorithm (BFPA) (Rodrigues et al., Citation2015) are used to examine the efficacy and efficiency of proposed BASO. Table outlines the detailed parameter settings of utilised algorithms. Note that the α and β of BASO are chosen according to (Zhao et al., Citation2019a). To ensure a fair comparison, the population size (N) and the maximum number of iterations (T) are fixed at 10 and 100, respectively.
Table 3. Parameter setting.
In this paper, six evaluation metrics include the best fitness, worst fitness, mean fitness, accuracy, feature size, and the computational time are used to measure the performance of proposed approaches. Each algorithm is repeated with M independent runs to achieve the statistical meaningful result. Finally, the average metrics obtained from M independent runs are presented as the experimental results. Note that the total number of function evaluations (NFE) for the proposed BASO is N × T × K × M. In this study, we set the K and M at 10 and 20. All the analysis is done in MATLAB 9.3 using a computer with Intel Core i5-9400 CPU 2.90 GHz and 16.0GB RAM.
5.3. Assessments of proposed BASO approaches in feature selection
In the first part of the experiment, the assessments on eight proposed BASO algorithms are performed. Tables , , and exhibit the best, worst and mean fitness values of proposed BASO approaches. Note that the best result for each approach is bolded. As can be observed, BASO-S1 and BASO-S2 scored the lowest best fitness values on most of the datasets (8 datasets), followed by BASO-S3 and BASO-V4 (6 datasets). Out of twenty-two datasets, BASO-S1 and BASO-V1 perceived the smallest worst fitness value in 5 datasets. As for mean fitness value, the best approach was found to be BASO-S1, which provided better performance than other approaches in feature selection.
Table 4. The best fitness value of proposed BASO approaches on 22 datasets.
Table 5. Result of worst fitness value of proposed BASO approaches on 22 datasets.
Table 6. Result of mean fitness value of proposed BASO approaches on 22 datasets.
Tables and outline the experimental results of accuracy and feature size of proposed BASO approaches. Inspecting the result in Table , BASO-S1 perceived the highest accuracy on 6 datasets, followed by BASO-S2 and BASO-V2 (4 datasets). Eventually, BASO-S1 outperformed other approaches in evaluating the significant features, thus leading to excellent results. In Table , BASO-S1 obtained the smallest number of selected features (feature size) on 9 datasets. In comparison with other approaches, BASO-S1 can usually provide a smaller feature size. Based on the results obtained, the BASO that contributed to the optimal performance was BASO-S1.
Table 7. Result of accuracy of proposed BASO approaches on 22 datasets.
Table 8. Result of feature size of proposed BASO approaches on 22 datasets.
Furthermore, the Wilcoxon signed-rank test with a 5% significant level (α=0.05) is used to examine whether the performance of BASO is significantly better than other algorithms. In this test, if the p-value is less than 0.05, then there is a significant difference between the performance of two algorithms; otherwise, the performances of the two algorithms are similar. Table outlines the p-value of the Wilcoxon signed-rank test for BASO-S1 and other approaches. In Table , the symbols of “w / t / l” indicate the proposed BASO-S1 was superior to (win), equal to (tie), and inferior to (lose) other algorithms. The result shows that the classification performances of BASO-S1 and BASO-S2 were similar. However, by making a one to one comparison (BASO-S1 versus BASO-S2), the algorithm that contributed to the optimal classification performance was found to be BASO-S1. From the point of view, BASO-S1 showed superior performance against other approaches in feature selection. Hence, only BASO with transfer function S1 is used in the rest of this paper.
Table 9. The p-value of Wilcoxon signed rank test for BASO-S1 and other BASO approaches.
5.4. Comparison with Other Algorithms
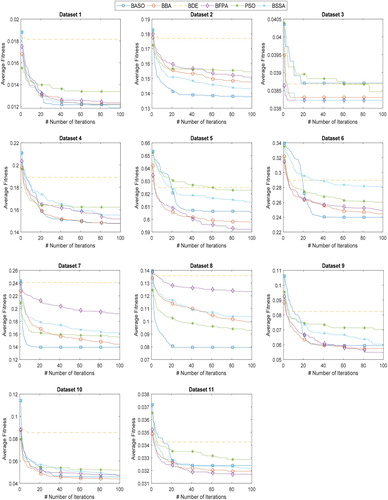
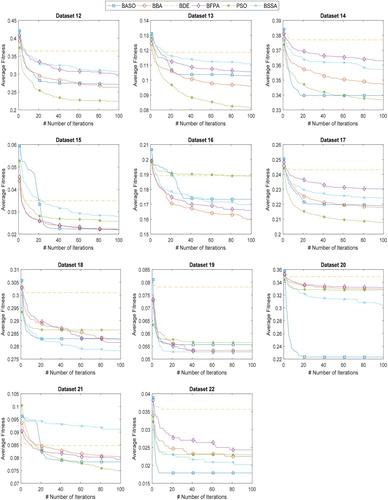
In the second part of the experiment, the performance of the proposed BASO is further compared with BBA, BDE, BFPA, PSO, and BSSA. Figure and Figure illustrate the convergence curves of proposed BASO and other algorithms on 22 datasets. From Figure and Figure , BASO contributed better convergence on most of the datasets, especially on dataset 2, 7, 8, 20, and 22. This finding indicates that BASO is highly capable of searching for an optimal solution, which leads to satisfactory performance. Taking dataset 22 (Leukemia) as an example, BASO converged faster than other algorithms to find the global optimum. In comparison with other algorithms, BASO offered a very high diversity, thus resulting in optimal fitness value.
Tables , , and show the experimental results of the best, worst, and mean fitness values of six different algorithms. In these Tables, the best result is highlighted with bold text. As can be seen, the performance of BASO was competitive against others. Among rivals, BASO achieved the optimal best and mean fitness values on 10 and 6 datasets, respectively. In terms of consistency, BFPA yielded the lowest STD of fitness value on most of the datasets, which showed high consistency in this work.
Table 10. Result of best fitness value of six different algorithms on 22 datasets.
Table 11. Result of worst fitness value of six different algorithms on 22 datasets.
Table 12. Result of mean fitness value of six different algorithms on 22 datasets.
Based on the results obtained, BASO has shown to be the best algorithm that contributed to the optimal performance in this work. The observed improvements in searching the global optimum (best solution) are attributed to the attractive force or repulsive force between the atom and its neighbour in BASO. Initially, the attractive force promotes exploration and enable the atoms to search globally. At the end of the iteration, the repulsive force encourages the atoms to search locally around the promising region. This, in turn, will make a well stable balance between global and local search, thus resulting in superior performance.
Table exhibits the result of the accuracy of six different algorithms on 22 datasets. As can be seen, the classification performance has shown great improvement when the feature selection algorithms are utilised. The results clearly show the effectiveness of feature selection in classification tasks. In Table , BASO contributed the highest accuracy on dataset 2, 4, 6, 7, 8, 15, 20, 21, and 22 (nine datasets). The second best algorithm was found to be BFPA (seven datasets), followed by BSSA (five datasets). Correspondingly, BASO overtakes other algorithms in evaluating the significant features from the large available feature set.
Table 13. Result of accuracy of six different algorithms on 22 datasets.
Table displays the result of feature size (number of selected features) of six different algorithms on 22 datasets. In terms of feature size, BASO can usually affirm the minimal number of selected features, followed by BSSA. Based on the result obtained, BASO gave the smallest feature size on 10 datasets. The results validate that BASO was good at eliminating the irrelevant and redundant features, thus, providing high prediction accuracy. Among rivals, BASO is highly capable of finding significant features, which can contribute to better classification performance.
Table 14. Result of feature size of six different algorithms on 22 datasets.
Table outlines the p-value of the Wilcoxon signed-rank test for BASO and its competitors. The experimental results clearly show that proposed BASO was a useful tool for feature selection problems. Furthermore, the result of computational time of six different algorithms is shown in Table . As can be observed, PSO was found to be the fastest feature selection algorithm, which contributes to the lowest computational time. Even though BASO cannot ensure the fastest processing speed, however, it can usually select a smaller number of significant features to achieve the highest classification accuracy.
Table 15. The p-value of Wilcoxon signed rank test for BASO and other algorithms.
Table 16. Result of computational time of six different algorithms on 22 datasets.
There are several limitations can be found within this research. First, the parameter settings of BASO are fixed in this work. For other applications, the users are recommended to test the parameters in order to get the best performance. Second, we only apply the KNN as the learning algorithm to compute the classification performance in feature selection. However, other popular and powerful learning algorithms such as support vector machine (SVM), convolutional neural network (CNN), and random forest (RF) can be implemented to boost the result.
6. Conclusion
In this paper, new binary variants of ASO, namely binary atom search optimisation (BASO), are proposed to solve the feature selection problem in classification tasks. Eight transfer functions (from S-shaped and V-shaped family) are used in this study to convert the continuous ASO into a binary version. Besides, a novel position updating rule is designed to enhance the performance of BASO in feature selection. Twenty-two datasets are utilised to validate the performance of the proposed BASO in feature selection. Our results indicated that BASO with S-shaped transfer function (S1) is highly capable of selecting the relevant features as compared to other BASO approaches. Moreover, the performance of the proposed BASO was further compared with BBA, BDE, BFPA, PSO, and BSSA. The experimental results proved the superiority of proposed BASO in terms of classification accuracy, feature size, and convergence speed. Future research can focus on the efficacy of BASO in the classification problem, as well as other binary optimisation tasks.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgement
The authors would like to thank the Skim Zamalah UTeM for supporting this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Al-Ani, A. (2005). Feature Subset Selection Using Ant Colony Optimization. https://opus.lib.uts.edu.au/handle/10453/6181
- Al-Tashi, Q., Kadir, S. J. A., Rais, H. M., Mirjalili, S., & Alhussian, H. (2019). Binary optimization using hybrid grey wolf optimization for feature selection. IEEE Access, 7, 39496–39508. https://doi.org/10.1109/ACCESS.2019.2906757
- Arora, S., & Anand, P. (2019). Binary butterfly optimization approaches for feature selection. Expert Systems with Applications, 116, 147–160. https://doi.org/10.1016/j.eswa.2018.08.051
- Chuang, L.-Y., Chang, H.-W., Tu, C.-J., & Yang, C.-H. (2008). Improved binary PSO for feature selection using gene expression data. Computational Biology and Chemistry, 32(1), 29–38. https://doi.org/10.1016/j.compbiolchem.2007.09.005
- Emary, E., Zawbaa, H. M., & Hassanien, A. E. (2016a). Binary grey wolf optimization approaches for feature selection. Neurocomputing, 172, 371–381. https://doi.org/10.1016/j.neucom.2015.06.083
- Emary, E., Zawbaa, H. M., & Hassanien, A. E. (2016b). Binary ant lion approaches for feature selection. Neurocomputing, 213, 54–65. https://doi.org/10.1016/j.neucom.2016.03.101
- Faris, H., Hassonah, M. A., Al-Zoubi, A. M., Mirjalili, S., & Aljarah, I. (2017). A multi-verse optimizer approach for feature selection and optimizing SVM parameters based on a robust system architecture. Neural Computing and Applications, 30(8), 2355–2369. https://doi.org/10.1007/s00521-016-2818-2
- Faris, H., Mafarja, M. M., Heidari, A. A., Aljarah, I., Al-Zoubi, A. M., Mirjalili, S., & Fujita, H. (2018). An efficient binary salp swarm algorithm with crossover scheme for feature selection problems. Knowledge-Based Systems, 154, 43–67. https://doi.org/10.1016/j.knosys.2018.05.009
- Huang, C.-L., & Wang, C.-J. (2006). A GA-based feature selection and parameters optimizationfor support vector machines. Expert Systems with Applications, 31(2), 231–240. https://doi.org/10.1016/j.eswa.2005.09.024
- Kennedy, J. (2011). Particle swarm Optimization. In Claude Sammut & Geoffrey I. Webb (Eds.), Encyclopedia of machine learning (pp. 760–766). Springer.https://doi.org/10.1007/978-0-387-30164-8_630.
- Kennedy, J., & Eberhart, R. C. (1997). A discrete binary version of the particle swarm algorithm. Computational Cybernetics and Simulation 1997 IEEE International Conference on Systems, Man, and Cybernetics, 5, 4104–4108. https://doi.org/10.1109/ICSMC.1997.637339
- Labani, M., Moradi, P., Ahmadizar, F., & Jalili, M. (2018). A novel multivariate filter method for feature selection in text classification problems. Engineering Applications of Artificial Intelligence, 70, 25–37. https://doi.org/10.1016/j.engappai.2017.12.014
- Liu, M., Xu, C., Luo, Y., Xu, C., Wen, Y., & Tao, D. (2018). Cost-Sensitive feature selection by Optimizing F-Measures. IEEE Transactions on Image Processing, 27(3), 1323–1335. https://doi.org/10.1109/TIP.2017.2781298
- Mafarja, M., Aljarah, I., Faris, H., Hammouri, A. I., Al-Zoubi, A. M., & Mirjalili, S. (2019). Binary grasshopper optimisation algorithm approaches for feature selection problems. Expert Systems with Applications, 117, 267–286. https://doi.org/10.1016/j.eswa.2018.09.015
- Mafarja, M., Aljarah, I., Heidari, A. A., Faris, H., Fournier-Viger, P., Li, X., & Mirjalili, S. (2018). Binary dragonfly optimization for feature selection using time-varying transfer functions. Knowledge-Based Systems, 161, 185–204. https://doi.org/10.1016/j.knosys.2018.08.003
- Mafarja, M. M., & Mirjalili, S. (2017). Hybrid Whale Optimization algorithm with simulated annealing for feature selection. Neurocomputing, 260, 302–312. https://doi.org/10.1016/j.neucom.2017.04.053
- Mirhosseini, M., & Nezamabadi-pour, H. (2018). BICA: A binary imperialist competitive algorithm and its application in CBIR systems. International Journal of Machine Learning and Cybernetics, 9(12), 2043–2057. https://doi.org/10.1007/s13042-017-0686-4
- Mirjalili, S., & Lewis, A. (2013). S-shaped versus V-shaped transfer functions for binary particle swarm Optimization. Swarm and Evolutionary Computation, 9, 1–14. https://doi.org/10.1016/j.swevo.2012.09.002
- Mirjalili, S., Mirjalili, S. M., & Yang, X.-S. (2014). Binary bat algorithm. Neural Computing and Applications, 25(3-4), 663–681. https://doi.org/10.1007/s00521-013-1525-5
- Pashaei, E., & Aydin, N. (2017). Binary black hole algorithm for feature selection and classification on biological data. Applied Soft Computing, 56, 94–106. https://doi.org/10.1016/j.asoc.2017.03.002
- Peng, H., Long, F., & Ding, C. (2005). Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence, 27(8), 1226–1238. https://doi.org/10.1109/TPAMI.2005.159
- Rashedi, E., Nezamabadi-pour, H., & Saryazdi, S. (2009). GSA: A gravitational search algorithm. Information Sciences, 179(13), 2232–2248. https://doi.org/10.1016/j.ins.2009.03.004
- Rashedi, E., Nezamabadi-pour, H., & Saryazdi, S. (2010). BGSA: Binary gravitational search algorithm. Natural Computing, 9(3), 727–745. https://doi.org/10.1007/s11047-009-9175-3
- Rodrigues, D., Pereira, L. A. M., Nakamura, R. Y. M., Costa, K. A. P., Yang, X.-S., Souza, A. N., & Papa, J. P. (2014). A wrapper approach for feature selection based on Bat algorithm and optimum-path forest. Expert Systems with Applications, 41(5), 2250–2258. https://doi.org/10.1016/j.eswa.2013.09.023
- Rodrigues, D., Yang, X.-S., Souza, A. N. d., & Papa, J. P. (2015). Binary flower pollination algorithm and Its application to feature selection. In X.-S. Yang (Ed.), Recent Advances in swarm Intelligence and Evolutionary Computation (pp. 85–100). Springer. https://doi.org/10.1007/978-3-319-13826-8_5
- Siddique, N., & Adeli, H. (2014). Water Drop algorithms. International Journal on Artificial Intelligence Tools, 23(06), 1430002. https://doi.org/10.1142/S0218213014300026
- Too, J., Abdullah, A. R., Mohd Saad, N., & Mohd Ali, N. (2018). Feature selection based on binary tree growth algorithm for the classification of Myoelectric Signals. Machines, 6(4), 65. https://doi.org/10.3390/machines6040065
- UCI Machine Learning Repository. (n.d.). Retrieved March 24, 2019, from https://archive.ics.uci.edu/ml/index.php
- Wang, C., Hu, Q., Wang, X., Chen, D., Qian, Y., & Dong, Z. (2018). Feature selection based on Neighborhood Discrimination Index. IEEE Transactions on Neural Networks and Learning Systems, 29(7), 1–14. https://doi.org/10.1109/TNNLS.2017.2710422 doi: 10.1109/TNNLS.2018.2845261
- Wang, M., Wan, Y., Ye, Z., & Lai, X. (2017). Remote sensing image classification based on the optimal support vector machine and modified binary coded ant colony optimization algorithm. Information Sciences, 402, 50–68. https://doi.org/10.1016/j.ins.2017.03.027
- Wolpert, D. H., & Macready, W. G. (1997). No free lunch theorems for optimization. IEEE Transactions on Evolutionary Computation, 1(1), 67–82. https://doi.org/10.1109/4235.585893
- Xue, B., Zhang, M., & Browne, W. N. (2014). Particle swarm optimisation for feature selection in classification: Novel initialisation and updating mechanisms. Applied Soft Computing, 18(Supplement C), 261–276. https://doi.org/10.1016/j.asoc.2013.09.018
- Zhao, W., Wang, L., & Zhang, Z. (2019a). Atom search optimization and its application to solve a hydrogeologic parameter estimation problem. Knowledge-Based Systems, 163, 283–304. https://doi.org/10.1016/j.knosys.2018.08.030
- Zhao, W., Wang, L., & Zhang, Z. (2019b). A novel atom search optimization for dispersion coefficient estimation in groundwater. Future Generation Computer Systems, 91, 601–610. https://doi.org/10.1016/j.future.2018.05.037
- Zorarpacı, E., & Özel, S. A. (2016). A hybrid approach of differential evolution and artificial bee colony for feature selection. Expert Systems with Applications, 62, 91–103. https://doi.org/10.1016/j.eswa.2016.06.004