Abstract
A portable immunochromatographic (IC) strip based on gold-labelled monoclonal antibodies was developed for the rapid detection of Pseudomonas syringae pv. maculicola, a plant pathogen. The IC strip detected 105 colony-forming units CFU/ml P. syringae pv. maculicola in pure culture within 10 min and had good specificity. Compared with sandwich enzyme-linked immunosorbent assay, the strip was equally sensitive; however, it is portable and time-efficient. On broccoli and radish seeds, the IC strips had a sensitivity of 5 × 105 CFU/ml and 106 CFU/ml, respectively. The results revealed that the IC strip could have potential applications for P. syringae pv. maculicola detection in plant seeds.
1. Introduction
Pseudomonas syringae pv. maculicola is a plant pathogen that causes irregular, brown and necrotic spots; water soaking on leaves; or pale rot on flowers (Cother & Noble, Citation2009; Keinath, Wechter, & Smith, Citation2006). This pathogen, which mainly infects cruciferae including cauliflower and broccoli (Zhao, Damicone, Demezas, Rangaswamy, & Bender, Citation2000), has a worldwide prevalence: Oklahoma and South Carolina (USA), Victoria (Australia), Loire Valley (France), Japan (Gironde & Manceau, Citation2012; Takikawa & Takahashi, Citation2014) and Hubei province (China).
Seeds represent an important transmission route of the pathogen (Chen et al., Citation2013). Therefore, an effective method to prevent pathogen transmission is through seed quarantines. Conventional methods used in the detection and identification of plant pathogens, including pathogen isolation with appropriate culture media and confirmation of pathogenicity, are both time-consuming and labour-intensive.
Rapid identification methods such as polymerase chain reaction (PCR), high-performance liquid chromatography (HPLC), enzyme-linked immunosorbent assay (ELISA) and strip tests have been used to detect plant pathogens. PCR has been used to detect P. syringae pv. actinidiae and P. syringae pv. tomato in kiwi fruits and tomatoes, respectively (Gallelli, Talocci, L’Aurora, & Loreti, Citation2012; Rees-George et al., Citation2010; Vanneste et al., Citation2011; Zaccardelli, Spasiano, Bazzi, & Merighi, Citation2005). HPLC and ELISA have been used to detect the presence of phytotoxin coronatine produced by P. syringae (Sreedharan, Penaloza-Vazquez, Escober, Bender, & Rayas-Duarte, Citation2009; Zhao et al., Citation2001). Gold-labelled chromatography strips are powerful tools for biological analyses. Strips have been widely used in the detection of chemicals, heavy metals, toxins and plant pathogens (Kuang, Xing et al., Citation2013; Liu et al., Citation2014; Wang et al., Citation2011; Zhao et al., Citation2011).
In this study, we developed a monoclonal antibody-based portable immunochromatographic (IC) strip for the detection of P. syringae pv. maculicola. The IC strip was evaluated in broccoli and radish seeds.
2. Materials and methods
2.1. Reagents and instruments
Hydrogen tetrachloroaurate hydrate, trisodium citrate, goat anti-mouse IgG antibody, Freund’s complete adjuvant and Freund’s incomplete adjuvant were purchased from Sigma–Aldrich (Shanghai, China). A low-flow nitrocellulose (NC) membrane (UniSart CN 95) and a glass fibre membrane were obtained from Whatman (Dassel, Germany). All reagents were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China). The CM4000 Guillotine Cutting Module and BioJet Quanti3000 dispenser used in this study was supplied by Bio Dot, Inc. (Irvine, CA, USA). The microplate reader was obtained from Thermo Scientific (MK3, Chicago, IL, USA).
2.2. Bacterial strains and culture conditions
P. syringae pv. maculicola (NCPPB 1820), Burkholderia glumae (NCPPB 3591), Xanthomonas oryzae pv. oryzicola (NCPPB 1585), Xanthomonas campestris pv. oryzae (NCPPB 3002), Pantoea stewartii subsp. stewartii (NCPPB 449) and P. syringae pv. syringae (NCPPB 2844) were kindly donated by Hunan Entry-Exit Inspection and Quarantine Bureau. B. glumae (NCPPB 3591) was cultured at 35°C in Brian Heart Infusion (BHI) medium; the other bacterial strains were cultured at 27°C in BHI medium.
2.3. Preparation of monoclonal antibodies (mAb) against P. syringae pv. maculicola
Boiled P. syringae pv. maculicola cells were used as immunogen. The immunization methodology was similar to that previously reported (Feng et al., Citation2013). The mouse with the highest titre against P. syringae pv. maculicola cells in indirect ELISA were sacrificed, and the spleen was fused with Sp2/0 murine myeloma cells. Positive cell lines against P. syringae pv. maculicola cells were selected by indirect ELISA and subcloned by the limited dilution method. mAb were produced in mice and purified with the caprylic acid-ammonium sulphate precipitation method. Sandwich ELISA of P. syringae pv. maculicola based on 1C3 as capture antibody and 6B6 as detection antibody was developed following a pairwise study (Kuang, Wang et al., Citation2013).
2.4. Preparation of colloidal gold- and gold nanoparticles-conjugated mAb
Colloidal gold (30 nm in diameter) was synthesized according to the method reported by Frens (Citation1973). Briefly, 300 ml of 0.01% HAuCl4 solution was thoroughly boiled under magnetic stirring; 4.8 ml of 1% trisodium citrate was quickly added to the solution and boiled for 10 min until the colour was wine-red. The diameter of the colloidal gold was confirmed by transmission electron microscopy. mAb 6B6 was conjugated to colloidal gold (Liu et al., Citation2014). Briefly, the pH value of colloidal gold solution (1 ml) was adjusted to 8 with 0.1 M K2CO3. Subsequently, mAb was added, reaching final concentrations of 2–10 μg/ml. Following a 2-h incubation, the solution was centrifuged (6000g for 20 min at 4°C). Unconjugated gold nanoparticles (GNPs) and mAb in the supernatant were discarded. Subsequently, GNP-labelled mAb was blocked with blocking buffer (PBS, 2% BSA, and 0.2% fucose) for 2 h. Lastly, the conjugate was washed and stored in 150-μl suspending buffer (PBS, 0.1% Tween 20 and 0.2% fucose).
2.5. IC strip
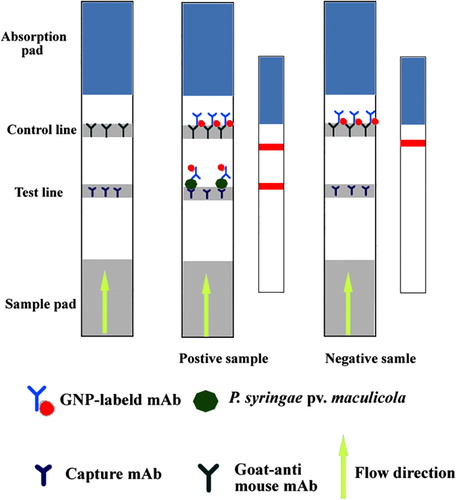
The preparation of the IC strip is shown in . The absorption pad, NC membrane and sample pad were pasted to the plastic backing board. Using the BioJet Quanti3000 dispenser, mAb 1C3 and goat anti-mouse IgG antibody (0.5 mg/ml) were sprayed on the NC membrane as test and control lines, respectively. The backing board was air-dried for 2 h at 37°C and cut into individual strips (4 mm wide). The strips were stored with desiccant at 4°C.
2.6. Evaluation of IC strip for P. syringae pv. maculicola
P. syringae pv. maculicola in pure culture were serially diluted with phosphate-buffered saline containing 0.1% Tween 20 (PBST) to concentrations that ranged from 108 CFU/ml to 103 CFU/ml. The specificity of the IC strip was evaluated with five plant bacteria, which were previously diluted to 108 CFU/ml. Before the IC strip test, 150 μl sample, 7 µl GNP-labelled mAb and 43 μl PBST were transferred to a 5-ml tube and allowed to react for 5 min. The sample pad of the strip was immersed in the solution and allowed to react for 10 min at 37°C. The results were either observed with the naked eye or assessed with a BioDot TSR3000 Membrane Strip reader (Gene Company Limited, Shanghai Branch, Shanghai, China). One hundred microlitres of each standard was added to the microplate and subjected to sandwich ELISA (Kuang, Wang et al., Citation2013).
2.7. Detection of seed samples with IC strip
Broccoli and radish seeds, which were obtained from Hunan Entry-Exit Inspection and Quarantine Bureau, were free of P. syringae pv. maculicola. P. syringae pv. maculicola-infected seed samples were prepared as previously described (Chen et al., Citation2013). Briefly, seeds were first disinfected with 3% NaClO for 10 min, washed three times with sterilized water and immersed overnight in PBS (500 seeds in 10 ml) at 4°C. Following centrifugation at 3000 rpm for 10 min, the supernatant was further centrifuged at 10,000 rpm for 10 min and the resulting sediment was discarded. The extract was spiked with cultured P. syringae pv. maculicola at 107, 2 × 106, 106, 2 × 105 and 105 CFU/ml. Extract solution was used as negative control. The samples were subjected to the IC strip and sandwich ELIA as described in 2.6.
3. Results and discussion
3.1. Principle and optimization of the strip
This IC strip detected P. syringae pv. maculicola. As shown in Figure 1, the samples were transferred from the sample pad to the absorption pad via capillary action. Positive samples (i.e., infected with P. syringae pv. maculicola) first reacted with the GNP-labelled mAb 6B6 and were captured by mAb 1C3 in the test line; excess GNP-labelled mAb was captured by the goat anti-mouse IgG antibody in the control line. Therefore, both test and control lines were red for positive samples. However, only the control line was red for negative samples.
To increase the sensitivity of the IC strip, concentrations of capture antibody 1C3 used in the test line and of 6B6 used for GNP conjugation were optimized. The optimal capture antibody concentration in this study was 6 mg/ml (4 µg in the test line), which was the original concentration. Lower concentrations reduced the colour–colour intensity of the test line, indicative of insufficient capture. Detection antibody concentration was 2 μg/ml in colloidal gold. Lower concentrations reduced the colour–colour intensity of the test line, because of inadequate GNP-labelled mAb. Higher concentrations increased the background of the test line, probably as a result of non-specific binding of excess GNP-labelled mAb (data not shown).
3.2. Evaluation of the IC strip for P. syringae pv. maculicola
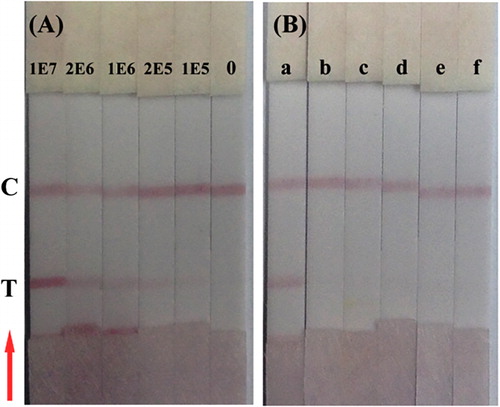
The sensitivity of the IC strip was assessed with the naked eye. As shown in , the bacterial concentrations on the test line increased from 105 CFU/ml to 107 CFU/ml, and the test line was blank for the negative control. Therefore, the sensitivity of the IC strip was 105 CFU/ml P. syringae pv. maculicola. The specificity results () revealed that the IC strip did not cross-react with P. syringae pv. syringae, B. glumae, X. oryzae pv. oryzicola, X. campestris pv. Oryzae or P. stewartii subsp. stewartii. No cross-reactivity with P. syringae pv. syringae suggests that the developed IC strip was specific to P. syringae pv. maculicola.
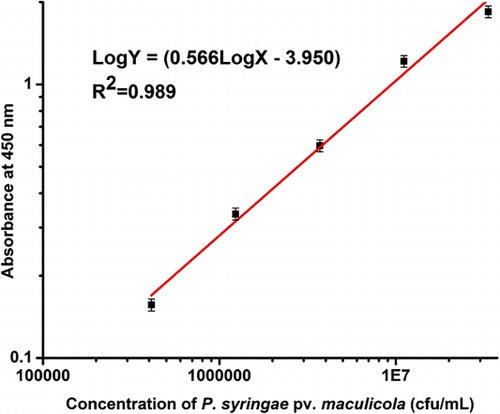
Sandwich ELISA detection, based on the same mAbs with IC strip, was performed. As shown in , the limit of detection (calculated as mean blank + 3SD) and working range of sandwich ELISA were 1.5 × 105 CFU/ml and 3 × 105 CFU/ml to 2 × 107 CFU/ml, respectively. shows the comparison of the IC strip with sandwich ELISA. The developed IC strip was as sensitive as sandwich ELISA; however, the IC strip is portable and faster.
Table 1. Comparison of IC strip with sandwich ELISA for P. syringae pv. maculicola.
3.3. Detection of P. syringae pv. maculicola on seed samples
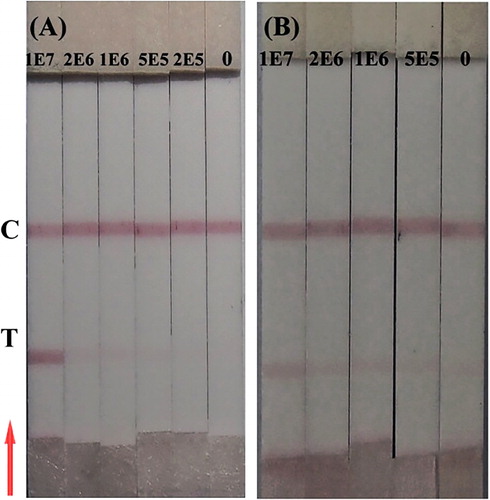
Two types of cruciferous vegetable seed samples (broccoli and radish seeds) were subjected to the developed IC strip. As shown in , the test lines were red when broccoli seed sample (with 5 × 105 CFU/ml P. syringae pv. maculicola) and the radish seed sample (with 1 × 106 CFU/ml P. syringae pv. maculicola) were subjected to the IC strip. Different detection limits indicate matrix effects.
4. Conclusion
An IC strip was developed and optimized for the detection of P. syringae pv. maculicola. The detection limits of the IC strip were 105 CFU/ml for pure culture and 5 × 105 CFU/ml for broccoli seeds. The results revealed that the IC strip was specific to P. syringae pv. maculicola, and faster than sandwich ELISA. The developed IC strip has potential applications in the detection of P. syringae pv. maculicola in plant seeds.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen, Y. P., Zou, M. Q., Wang, D. N., Li, Y. L., Xue, Q., Xie, M. X., & Qi, C. (2013). An immunosensor based on magnetic relaxation switch and polystyrene microparticle-induced immune multivalency enrichment system for the detection of Pantoea stewartii subsp. stewartii. Biosensors and Bioelectronics, 43, 6–11. doi:10.1016/j.bios.2012.11.022
- Cother, E., & Noble, D. (2009). Identification of blossom blight in stock (Matthiola incana) caused by Pseudomonas syringae pv. maculicola. Australasian Plant Pathology, 38, 242–246.
- Feng, M., Yong, Q., Wang, W., Kuang, H., Wang, L., & Xu, C. (2013). Development of a monoclonal antibody-based ELISA to detect Escherichia coli O157:h77. Food and Agricultural Immunology, 24, 481–487. doi:10.1080/09540105.2012.716026
- Frens, G. (1973). Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature, 241(105), 20–22.
- Gallelli, A., Talocci, S., L’Aurora, A., & Loreti, S. (2012). Detection of Pseudomonas syringae pv. actinidiae, causal agent of bacterial canker of kiwifruit, from symptomless fruits and twigs, and from pollen. Phytopathologia Mediterranea, 50, 462–472.
- Gironde, S., & Manceau, C. (2012). Housekeeping gene sequencing and multilocus variable-number tandem-repeat analysis to identify subpopulations within Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. tomato that correlate with host specificity. Applied and Environmental Microbiology, 78, 3266–3279. doi:10.1128/AEM.06655-11
- Keinath, A. P., Wechter, W. P., & Smith, J.P. (2006). First report of bacterial leaf spot on leafy Brassica greens caused by Pseudomonas syringae pv. maculicola in South Carolina. Plant Disease, 90, 683–683. doi:10.1094/PD-90-0683C
- Kuang, H., Wang, W., Xu, L., Ma, W., Liu, L., Wang, L., & Xu, C. (2013). Monoclonal antibody-based sandwich ELISA for the detection of staphylococcal enterotoxin A. International Journal of Environmental Research and Public Health, 10, 1598–1608. doi:10.3390/ijerph10041598
- Kuang, H., Xing, C., Hao, C., Liu, L., Wang, L., & Xu, C. (2013). Rapid and highly sensitive detection of lead ions in drinking water based on a strip immunosensor. Sensors, 13, 4214–4224. doi:10.3390/s130404214
- Liu, L., Luo, L., Suryoprabowo, S., Peng, J., Kuang, H., & Xu, C. (2014). Development of an immunochromatographic strip test for rapid detection of ciprofloxacin in milk samples. Sensors, 14, 16785–16798.
- Rees-George, J., Vanneste, J. L., Cornish, D. A., Pushparajah, I. P. S., Yu, J., Templeton, M. D., & Everett, K. R. (2010). Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathology, 59, 453–464. doi:10.1111/j.1365-3059.2010.02259.x
- Sreedharan, A., Penaloza-Vazquez, A., Escober, M. C., Bender, C. L., & Rayas-Duarte, P. (2009). Simple and rapid capillary zone electrophoresis method for the detection of coronamic acid, a precursor to the Pseudomonas syringae phytotoxin coronatine. Journal of Agricultural and Food Chemistry, 57, 10518–10523. doi:10.1021/jf9024008
- Takikawa, Y., & Takahashi, F. (2014). Bacterial leaf spot and blight of crucifer plants (Brassicaceae) caused by Pseudomonas syringae pv. maculicola and P. cannabina pv. alisalensis. Journal of General Plant Pathology, 80, 466–474. doi:10.1007/s10327-014-0540-4
- Vanneste, J., Giovanardi, D., Yu, J., Cornish, D., Kay, C., Spinelli, F., & Stefani, E., (2011). Detection of Pseudomonas syringae pv. actinidiae in kiwifruit pollen samples. New Zealand Plant Protection, 64, 246–251.
- Wang, L., Chen, W., Ma, W., Liu, L., Ma, W., Zhao, Y., … Xu, C. (2011). Fluorescent strip sensor for rapid determination of toxins. Chemical Communications, 47, 1574–1576. doi:10.1039/C0CC04032K
- Zaccardelli, M., Spasiano, A., Bazzi, C., & Merighi, M. (2005). Identification and in planta detection of Pseudomonas syringae pv. tomato using PCR amplification of hrpZPst. European Journal of Plant Pathology, 111(1), 85–90. doi:10.1007/s10658-004-2734-7
- Zhao, W., Lu, J., Ma, W., Xu, C., Kuang, H., & Zhu S. (2011). Rapid on-site detection of Acidovorax avenae subsp. citrulli by gold-labeled DNA strip sensor. Biosensors and Bioelectronics, 26, 4241–4244. doi:10.1016/j.bios.2011.04.004
- Zhao, Y., Damicone, J. P., Demezas, D. H., Rangaswamy, V., & Bender, C. L. (2000). Bacterial leaf spot of leafy crucifers in Oklahoma caused by Pseudomonas syringae pv. maculicola. Plant Disease, 84, 1015–1020. doi:10.1094/PDIS.2000.84.9.1015
- Zhao, Y. F., Jones, W. T., Sutherland, P., Palmer, D. A., Mitchell, R. E., Reynolds, P. H. S., … Bender, C. L. (2001). Detection of the phytotoxin coronatine by ELISA and localization in infected plant tissue. Physiological and Molecular Plant Pathology, 58, 247–258. doi:10.1006/pmpp.2001.0334