ABSTRACT
An ultrasensitive and selective method for Bisphenol A (BPA) detection using aptamer-BPA and real-time quantitative polymerase chain reaction (RT-qPCR) technology is reported, in which biotin-modified aptamer DNA (BPA) is fixed on the inner wall of a streptavidin-coated PCR tube via biotin–avidin interactions and the template is fixed through the complementary base pairing with aptamer DNA. Upon the addition of BPA, the template DNA could be off from the inner wall of PCR tube, which results from aptamer DNA interacted specifically with BPA. Thus, the number of template DNA change result from the BPA concentration is readily seen by cycle threshold (Ct) values. The sensor exhibited an excellent linear response between Ct values and BPA concentration in the range of 1–500 nM and the detection limit of this method for aqueous BPA was as low as 0.7 nM. In addition, this method showed good selectivity for BPA over its analogues.
1. Introduction
2,2-bis(4-hydroxyphenyl)propane (Bisphenol A, BPA) is an all-important chemical raw material (Staples, Woodburn, Caspers, Hall, & Kleĉka, Citation2002), and mainly used for the production of polycarbonate plastics and epoxy resins (Willhite, Ball, & McLellan, Citation2008). Polycarbonate plastics and epoxy resins are widely produced in large quantities for use in the manufacturing of food contact materials, such as tableware, water pipes, baby bottles, food and drink packaging (Crain et al., Citation2007). Therefore, human exposure to BPA is inevitable in daily life. BPA is currently regarded as an environmental endocrine disrupting chemical (EDC) which can interfere with the endocrine system. Many studies strongly prove that BPA is harmful to the human body, which can cause the increasing rates of breast cancer, the decreasing sperm count in males, the changing of the uterus and ovaries, as well as the changing in neural and behavioural of infants and children (Carlsen, Giwercman, Keiding, & Skakkebæk, Citation1995; Long et al., Citation2000; Markey, Wadia, Rubin, Sonnenschein, & Soto, Citation2005; Newbold, Jefferson, & Padilla-Banks, Citation2007). Thus, rapid, simple and accurate detection of BPA in food related samples is of great importance for assuring human health. In particular, monitoring the BPA concentration in drinking water, is significant for food safety
Recently, various traditional instrument-based detection methods, such as capillary electrophoresis(CE) (Zhong, Tan, Ge, Wang, & Chen, Citation2011), high-performance liquid chromatography (HPLC) (Tan, Song, Wei, & Yi, Citation2012), liquid chromatography/mass spectrometry (LC/MS), gas chromatography/mass spectrometry (GC/MS) (Mudiam et al., Citation2011), and enzyme-linked immunosorbent assay (ELISA)+1 are frequently used for detection of BPA and its analogue (Yu, Liu, Song, Kuang, & Xu, Citation2016). Although these methods are sensitive and highly specific, they also require sample pre-treatment, expensive instruments as well as skilful operators. Electrochemical methods (Wang, Yang, & Wu, Citation2009; Yin et al., Citation2010), and fluorescence methods (Wang, Zeng, Zhao, & Lin, Citation2006), have also adopted for BPA detection due to the high sensitivity and cost-effective of these techniques. Determinations with electrochemical methods, however, the detection probe are usually immobilised to the electrode surface, resulting in a complicated detection process. As for fluorescence methods, the production of back ground signals derived from fluorophores and reaction mixtures could be interface the result. Hence, developing simple, sensitive, highly specific for detection of BPA is a great necessity.
DNA aptamers are distinctive structures composed of nucleotides or peptides to bind to a desired target ligand (Proske, Blank, Buhmann, & Resch, Citation2005). Under the action of the target, the DNA aptamers will produce conformational changing (Ellington & Szostak, Citation1990; Tuerk & Gold, Citation1990). Compared to the antibody, aptamers have some advantages such as thermal stability, reusability, and easy chemical synthesis, they have been widely used to recognise a wide range of substances from ions to cells in the analytical field (McKeague et al., Citation2014; Long, Zhu, & Wang, Citation2014; Wang et al., Citation2014; Wang et al., Citation2015; Xie et al., Citation2014; Zhu et al., Citation2015). Therefore, anti BPA aptamers were utilised in our developed method.
Real-time quantitative polymerase chain reaction (RT-qPCR) has become the most powerful tool in the field of science, medicine and diagnosis. RT-qPCR has been normally used for detecting for specific DNA fragments. The amount of specific DNA fragments are increasing exponentially during RT-qPCR, the fluorescent signal produced has been used as a quantitative indicator for the detection of specific DNA fragments (Kubista et al., Citation2006; Mestdagh et al., Citation2008; Schmittgen & Livak, Citation2008). As the amplification effect and quantitative measurement of RT-qPCR, it has the advantages of short operation time, high detection limit and favourable reproducibility (Adler, Wacker, & Niemeyer, Citation2003; Csordas et al., Citation2010; Jiang et al., Citation2012; KiatáHo & HanáAng, Citation2012; Kim, McCoy, Gee, González-Sapienza, & Hammock, Citation2010). Besides, the RT-qPCR technology can realise high throughput samples screening. It is suitable for the increasing testing requirements.
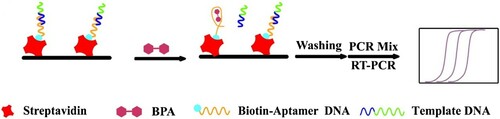
Herein, we reported a novel type of biosensor based on an aptamer specific to BPA for the highly sensitive and selective detection of BPA. The whole operation process was carried out in the PCR tube. In the first, the aptamer DNA and the template DNA were fixed to the PCR tube wall by specific binding force between biotin and streptavidin. Then the template DNA would detach from the aptamer DNA result from the aptamer DNA conformational changing with BPA. The rest template DNA was used as the template for the PCR amplification and the template DNA concentration would be reflected in the Ct values. At last, the Ct values as the indicator indirect reflected the target concentration. This novel sensor could be a facile, high-throughput and highly sensitive method for detection of BPA.
2. Materials and methods
2.1. Materials and reagents
2, 2-bis (4-hydroxyphenyl) propane (Bisphenol A, BPA) and its analogues were obtained from J&K Chemical Company, Shanghai. All oligonucleotides and Streptavidin were obtained from Sangon Biotech Co., Ltd. (Shanghai, China). iTaqTM Universal SYBR® Green Supermix was purchased from BIO-RAD (Shanghai, China). The water used in all experiments was deionised and purified to 18.2 MW (Millipore). All the other reagents were used as received without further purification. The sequences of DNA fragments were as follows:
2.2. RT-qPCR assay for BPA detection
Firstly, Each PCR tube was incubated with 20 μL of 0.8% glutaraldehyde solution at 37°C for 5 h in order to enhance their absorbability. Then the PCR tubes were treated with 20 μL streptavidin dissolved in 0.01 M carbonate buffer solution at 37°C for 2 h after washing three times with ultrapure water. The tubes were subsequently washed with PBST (10 mM PBS, pH 7.2, 0.05% Tween-20). After washing, the mixtures of template DNA and aptamer DNA were added, and the sample was incubated at 37°C for 40 min. The aptamer DNA was fixed on the surface of the PCR tubes by specific binding force between biotin and streptavidin. The DNA fragments which were not fixed would be cleaned by the hybridisation buffer (750 mM NaCl, 75 mM C6H5Na3O7, pH 8.0). And then various concentrations of BPA were added to each tube, incubated at 37°C for 40 min. Before testing, the DNA fragments in each tube were removed by washing three times with the hybridisation buffer other than the oligonucleotides which were fixed the surface of the PCR tubes.
The whole operation process was carried out in the PCR tubes. RT-qPCR was performed using a CFX-96 real-time PCR detection system. Before the PCR process, 20 μL PCR mixtures were added into each PCR tube. The 20 μL PCR mixtures were composed of upstream and downstream primer (each 2 μL, 500 nM), iTaqTM Universal SYBR® Green Supermix (10 μL), and ultrapure water (6 μL). The PCR cycling parameters consisted of an initial denaturation for 30 s at 95°C, followed by 39 cycles of denaturation for 5 s at 95°C and annealing for 30 s at 59°C. Fluorescence measurements were taken after each annealing step. A melting curve was obtained from 65°C to 95°C to detect potential nonspecific products, with the signal measured every 0.5°C.
2.3. Specificity analysis
Three types of BPA analogues Bisphenol C (BPC), Biphenolic acid (DPA), and Bisphenol B (BPB) were selected to evaluate the specificity of this assay. BPA and its three analogues were added at a concentration of 500 nM individually. All other procedures were identical to those mentioned in Section 2.2.
2.4. Recovery in water samples
To evaluate the practical analytical performance of the new sensor for the detection of BPA, we used tap water as a real sample. Tap water samples were spiked with BPA (1, 10, 50, and 100 nM). The spiked tap water samples were tested using our developed RT-qPCR based method.
3. Results and discussion
3.1. The principle of an aptamer-labeled sensor for ultrasensitive detection of BPA
The sensing process of the aptasensor for BPA detection is illustrated in Scheme 1. Template DNA and aptamer DNA were fixed the surface of the PCR tubes which had been treated by streptavidin. The aptamer DNA was immobilised by means of the specific binding force between biotin and streptavidin, the template DNA was hybridised with the aptamer DNA through the partly complementary base pairing. Thereafter the different concentrations of BPA solutions were added and then incubated for 40 min at 37°C. The aptamer DNA's structural has changed in the presence of BPA and this led to the template DNA detach from the surface of the PCR tubes. After removing the free DNA fragments, the PCR mixture was added for the PCR amplification and the initial template DNA concentration would be reflected in the Ct values. At last, the Ct values as the indicator indirect reflected the BPA concentration.
3.2. Optimisation of the sensing conditions
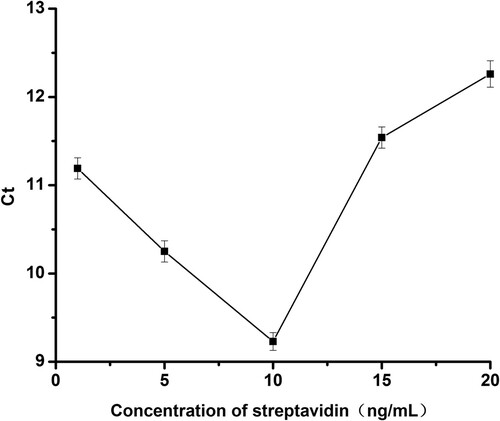
In this sensing system, several factors that may affect the quantitative analysis of BPA were optimised, including the concentration of streptavidin and the time for reacting between BPA and aptamer DNA. Streptavidin was used to fix the DNA fragment to the PCR tube wall via the biotin–avidin specific binding. Therefore, the concentration of streptavidin was one of the very important factors in measuring BPA concentration. As shown in , the Ct values responsed of the novel sensor based on different concentrations of streptavidin. The Ct values decreased with increasing the concentration of streptavidin up to 10 ng/mL and increased when the concentration of streptavidin exceeds 10 ng/mL. The minimum Ct value occurred when the concentration of streptavidin was 10 ng/mL. In other words, this new sensor was the most sensitive at this time. The streptavidin concentration of 10 ng/mL was also selected in the following experiments.
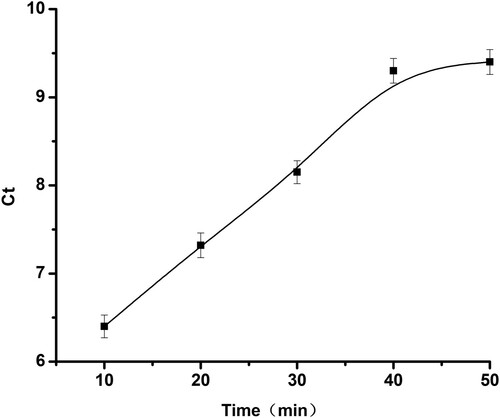
The effect of the time for reacting between BPA and aptamer DNA was also investigated. The time for reacting between BPA and aptamer DNA was a significant factor affecting the experimental sensitivity, which was determining whether aptamer DNA and BPA reacting completely. The reaction time was optimised when the streptavidin was the optimal concentration, and the results obtained were shown in . With the increase of reaction time, the Ct value also increased until 40 min. The Ct value was almost constant when the reaction time exceeds 40 min. At this point, it was means BPA and aptamer DNA reacted completely with the shortest time. Therefore, 40 min was selected as the time for reacting between BPA and aptamer DNA.
3.3. Sensitivity and selectivity of BPA detection
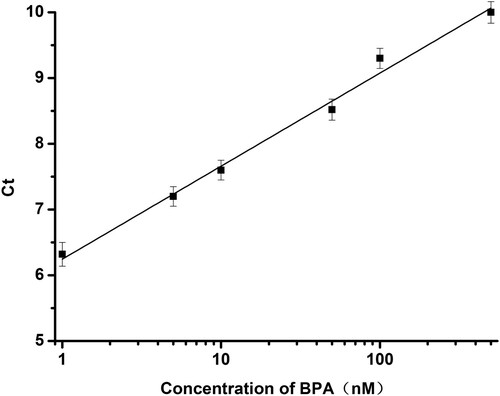
Under the optimum conditions described above, this method was initially applied to the analysis of BPA in water ample. In presence of diluted BPA standards in the modified PCR tubes, aptamer DNA conformational changed resulting in the decreasing of the template DNA. After BPA/aptamer interaction and the following PCR process, the amplification curves for each sample were collected using a fluorescence PCR instrument. As shown in , amplification curve was obtained by adding different concentrations of BPA. Since different amount of template DNA falls off by the addition of different concentration of BPA (0.5, 1, 5, 10, 50, 100, 500, 1000 nM), the concentration of template DNA using for template amplification was varied. The Ct values were increasing with increasing the concentration of BPA (). A good linear relationship between Ct values and BPA concentration was observed in the range of 1–500 nM BPA (), and the linear correlation equation was determined to be Ct = 6.25 + 1.41 lgC, where C is the concentration of BPA in nM. The limit of detection (LOD) was as low as 0.7 nM and the LOD was calculated as LOD = 3 (SD/S), where SD is the standard deviation of the response and S is the slope of the calibration curve.
Figure 3. The amplification curves at different concentrations of BPA in the range of 0.5 to 1000 nM. The addition of different concentration of BPA (0.5, 1, 5, 10, 50, 100, 500, 1000 nM).
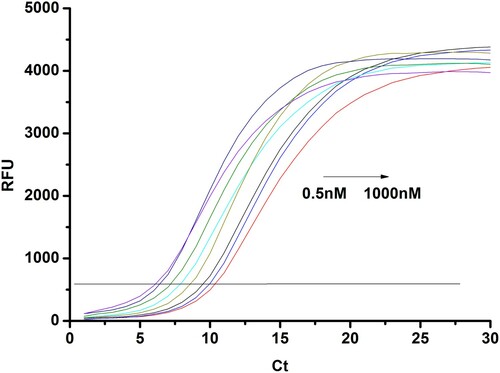
Figure 4. Ct value change for the detection of BPA at different concentrations. (0.5, 1, 5, 10, 50, 100, 500, 1000 nM).
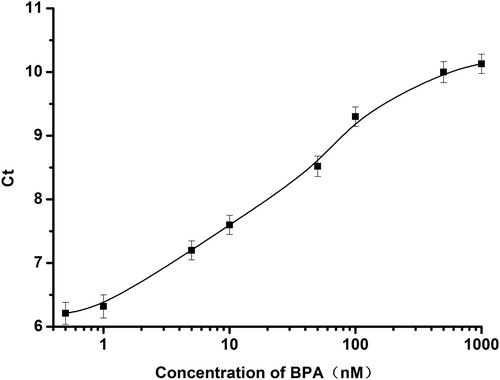
Figure 5. The linear relationship between the BPA concentration and the Ct value in the range of 1 to 500 nM.
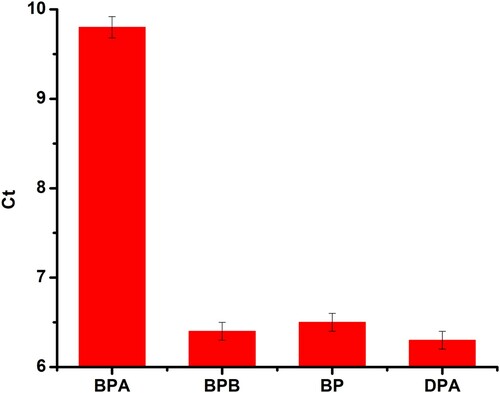
To evaluate selectivity of the aptasensor, BPA analogues, including Bisphenol B (BPB), Bisphenol C (BPC), Bophenolic acid (DPA), were tested at a concentration of 500 nM. The results were shown in , the Ct values were not obvious variation except for BPA, which demonstrated that these three BPA analogues would not recognise with the aptamer DNA. In other words, these three BPA analogues would not affect the detection of BPA by this method. Therefore, the novel aptasensor based on PCR for detecting BPA exhibited good specificity.
3.4. Detection of BPA in tap water samples
In order to examine the practicality of the developed sensor, the sensing system was applied to the detection of BPA in the tap water. Different amount of BPA standard solution was spiked to the tap water, making its final concentration respectively 1, 10, 50, 100 nM. The test results were shown in , the recoveries of detecting BPA in real samples were in the range of 96.8–104.0% with a RSD ranging from 2.75% to 4.74%. These satisfactory results demonstrated that this novel sensor could be acceptable for BPA detection in practical samples.
Table 1. Recovery experiments of BPA in tap water samples.
4. Conclusions
In our present study, we designed the novel sensor, using the Ct values as an indicator, based on BPA aptamer and its complementary for BPA detection. Ct values depended on the amount of the template DNA concentration, which was consistent with BPA concentration. Under optimal conditions, LOD of 0.7 nM was obtained in the range of 1 to 500 nM. In addition to high sensitivity, the aptamer-based method exhibited good specificity than other BPA analogues. The performance of the aptamer sensor evaluated in spiked tap water samples showed good recovery, which was in the range from 96.8% to 104.0% with a RSD ranging from 2.75% to 4.74%. This new sensor is credible and has enormous potential to detection of BPA in real water samples.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Adler, M., Wacker, R., & Niemeyer, C. M. (2003). A real-time immuno-PCR assay for routine ultrasensitive quantification of proteins. Biochemical and Biophysical Research Communications, 308, 240–250. doi: https://doi.org/10.1016/S0006-291X(03)01364-0
- Carlsen, E., Giwercman, A., Keiding, N., & Skakkebæk, N. E. (1995). Declining semen quality and increasing incidence of testicular cancer: Is there a common cause? Environmental Health Perspectives, 103, 137–139. doi: https://doi.org/10.1289/ehp.95103s7137
- Crain, D. A., Eriksen, M., Iguchi, T., Jobling, S., Laufer, H., LeBlanc, G. A., & Guillette, L. J., Jr. (2007). An ecological assessment of Bisphenol-A: Evidence from comparative biology. Reproductive Toxicology, 24, 225–239. doi: https://doi.org/10.1016/j.reprotox.2007.05.008
- Csordas, A., Gerdon, A. E., Adams, J. D., Qian, J., Oh, S. S., Xiao, Y., & Soh, H. T. (2010). Detection of proteins in serum by micromagnetic aptamer PCR (MAP) technology. Angewandte Chemie International Edition, 49, 355–358. doi: https://doi.org/10.1002/anie.200904846
- Ellington, A. D., & Szostak, J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. doi: https://doi.org/10.1038/346818a0
- Jiang, X., Cheng, S., Chen, W., Wang, L., Shi, F., & Zhu, C. (2012). Comparison of oligonucleotide-labeled antibody probe assays for prostate-specific antigen detection. Analytical Biochemistry, 424, 1–7. doi: https://doi.org/10.1016/j.ab.2012.02.004
- KiatáHo, H., & HanáAng, W. (2012). Immobilisation of quantum dots by bio-orthogonal PCR amplification and labelling for direct gene detection and quantitation. Chemical Communications, 48, 5467–5469. doi: https://doi.org/10.1039/c2cc30680h
- Kim, H.-J., McCoy, M., Gee, S. J., González-Sapienza, G. G., & Hammock, B. D. (2010). Noncompetitive phage anti-immunocomplex real-time polymerase chain reaction for sensitive detection of small molecules. Analytical Chemistry, 83, 246–253. doi: https://doi.org/10.1021/ac102353z
- Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., … Strömbom, L. (2006). The real-time polymerase chain reaction. Molecular Aspects of Medicine, 27, 95–125. doi: https://doi.org/10.1016/j.mam.2005.12.007
- Long, X., Steinmetz, R., Ben-Jonathan, N., Caperell-Grant, A., Young, P., Nephew, K. P., & Bigsby, R. M. (2000). Strain differences in vaginal responses to the xenoestrogen bisphenol A. Environmental Health Perspectives, 108, 243–247. doi: https://doi.org/10.1289/ehp.00108243
- Long, F., Zhu, A., & Wang, H. C. (2014). Optofluidics-based DNA structure-competitive aptasensor for rapid on-site detection of lead(II) in an aquatic environment. Analytica Chimica Acta, 849, 43–49. doi: https://doi.org/10.1016/j.aca.2014.08.015
- Markey, C. M., Wadia, P. R., Rubin, B. S., Sonnenschein, C., & Soto, A. M. (2005). Long-term effects of fetal exposure to low doses of the xenoestrogen bisphenol-A in the female mouse genital tract. Biology of Reproduction, 72, 1344–1351. doi: https://doi.org/10.1095/biolreprod.104.036301
- McKeague, M., Velu, R., Hill, K., Bardoczy, V., Meszaros, T., & DeRosa, M. C. (2014). Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins, 6, 2435–2452. doi: https://doi.org/10.3390/toxins6082435
- Mestdagh, P., Feys, T., Bernard, N., Guenther, S., Chen, C., Speleman, F., & Vandesompele, J. (2008). High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Research, 36, e143–e143. doi: https://doi.org/10.1093/nar/gkn725
- Mudiam, M. K. R., Jain, R., Dua, V. K., Singh, A. K., Sharma, V., & Murthy, R. (2011). Application of ethyl chloroformate derivatization for solid-phase microextraction–gas chromatography–mass spectrometric determination of bisphenol-A in water and milk samples. Analytical and Bioanalytical Chemistry, 401, 1695–1701. doi: https://doi.org/10.1007/s00216-011-5226-6
- Newbold, R. R., Jefferson, W. N., & Padilla-Banks, E. (2007). Long-term adverse effects of neonatal exposure to bisphenol A on the murine female reproductive tract. Reproductive Toxicology, 24, 253–258. doi: https://doi.org/10.1016/j.reprotox.2007.07.006
- Proske, D., Blank, M., Buhmann, R., & Resch, A. (2005). Aptamers – basic research, drug development, and clinical applications. Applied Microbiology and Biotechnology, 69, 367–374. doi: https://doi.org/10.1007/s00253-005-0193-5
- Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 3, 1101–1108. doi: https://doi.org/10.1038/nprot.2008.73
- Staples, C. A., Woodburn, K., Caspers, N., Hall, A. T., & Kleĉka, G. M. (2002). A weight of evidence approach to the aquatic hazard assessment of bisphenoi A. Human and Ecological Risk Assessment, 8, 1083–1105. doi: https://doi.org/10.1080/1080-700291905837
- Tan, X.-W., Song, Y.-X., Wei, R.-P., & Yi, G.-Y. (2012). Determination of trace bisphenol A in water using three-phase hollow fiber liquid-phase microextraction coupled with high performance liquid chromatography. Chinese Journal of Analytical Chemistry, 40, 1409–1414. doi: https://doi.org/10.1016/S1872-2040(11)60574-4
- Tuerk, C., & Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. doi: https://doi.org/10.1126/science.2200121
- Wang, K. Y., Liao, J., Yang, X. Y., Zhao, M., Chen, M., Yao, W. R., … Lan, X. P. (2015). A label-free aptasensor for highly sensitive detection of ATP and thrombin based on metal-enhanced PicoGreen fluorescence. Biosensors and Bioelectronics, 63, 172–177. doi: https://doi.org/10.1016/j.bios.2014.07.022
- Wang, J. C., Wang, Y. S., Xue, J. H., Zhou, B., Qian, Q. M., Wang, Y. S., … Liu, S. D. (2014). An ultrasensitive label-free assay of 8-hydroxy-2'-deoxyguanosine based on the conformational switching of aptamer. Biosensors and Bioelectronics, 58, 22–26. doi: https://doi.org/10.1016/j.bios.2014.02.047
- Wang, F., Yang, J., & Wu, K. (2009). Mesoporous silica-based electrochemical sensor for sensitive determination of environmental hormone bisphenol A. Analytica Chimica Acta, 638, 23–28. doi: https://doi.org/10.1016/j.aca.2009.02.013
- Wang, X., Zeng, H., Zhao, L., & Lin, J.-M. (2006). Selective determination of bisphenol A (BPA) in water by a reversible fluorescence sensor using pyrene/dimethyl β-cyclodextrin complex. Analytica Chimica Acta, 556, 313–318. doi: https://doi.org/10.1016/j.aca.2005.09.060
- Willhite, C. C., Ball, G. L., & McLellan, C. J. (2008). Derivation of a bisphenol A oral reference dose (RfD) and drinking-water equivalent concentration. Journal of Toxicology and Environmental Health, Part B, 11, 69–146. doi: https://doi.org/10.1080/10937400701724303
- Xie, Q., Tan, Y. Y., Guo, Q. P., Wang, K. M., Yuan, B. Y., Wan, J., & Zhao, X. Y. (2014). A fluorescent aptasensor for sensitive detection of human hepatocellular carcinoma SMMC-7721 cells based on graphene oxide. Anal Methods, 6, 6809–6814. doi: https://doi.org/10.1039/C4AY01213E
- Yin, H., Zhou, Y., Ai, S., Chen, Q., Zhu, X., Liu, X., & Zhu, L. (2010). Sensitivity and selectivity determination of BPA in real water samples using PAMAM dendrimer and CoTe quantum dots modified glassy carbon electrode. Journal of Hazardous Materials, 174, 236–243. doi: https://doi.org/10.1016/j.jhazmat.2009.09.041
- Yu, L., Liu, L.-Q., Song, S.-S., Kuang, H., & Xu, C.-L. (2016). Development of an immunochromatographic test strip and ic-ELISA for tetrabromobisphenol: A detection in lake water and rice pudding samples. Food and Agricultural Immunology, 27, 460–470. doi: https://doi.org/10.1080/09540105.2015.1126234
- Zhong, S., Tan, S. N., Ge, L., Wang, W., & Chen, J. (2011). Determination of bisphenol A and naphthols in river water samples by capillary zone electrophoresis after cloud point extraction. Talanta, 85, 488–492. doi: https://doi.org/10.1016/j.talanta.2011.04.009
- Zhu, Y.-Y., Deng, D.-Q., Xu, L.-G., Zhu, Y.-B., Wang, L.-M., Qi, B., & Xu, C.-L. (2015). Mercury-DNA interaction based detection of mercury ions by DNA amplification with high sensitivity and selectivity. Food and Agricultural Immunology, 26(4), 512–520. doi: https://doi.org/10.1080/09540105.2014.988127