ABSTRACT
The objectives of this study were to determine the antimicrobial susceptibility, phylotype, and virulence genes of Escherichia coli (E. coli) from cows with clinical mastitis. A total of 497 mastitis samples were collected and 92 E. coli isolates were identified. E. coli isolates were highly resistance to ampicillin (41.3%), followed by piperacillin (36.9%) and tetracycline (34.7%). Five intimin types, namely μR-ι2, λ, ξR/β2, νR-ε2, η, were observed among 75 EPEC strains with eae gene, with all the intimins were first detected in bovine mastitis milk. And 65 E. coli strains (70.7%) belonged to seven different O serotypes (O121, O91, O22, O26, O128, O111, and O113). For the phylogroup assignment, only 87 E. coli isolates could be assigned into phylogroup B1, A, C, and E. This study suggested the prevailing among EPEC strains isolated from mastitis in five provinces was important to understand the etiology of E. coli.
1. Introduction
Bovine mastitis, which affects dairy farms throughout the world, reduces milk production, negatively impacts milk quality, and represents a source of contamination for raw milk and dairy products (Halasa, Huijps, Østerås, & Hogeveen, Citation2007). Several bacteria cause mastitis in dairy cows, including Escherichia coli (Olde Riekerink, Barkema, Scholl, Poole, & Kelton, Citation2010), one of the most predominant pathogens(Zhang et al., Citation2018), Staphylococcus aureus (Olde Riekerink, Barkema, Kelton, & Scholl, Citation2008), and Streptococcus uberis(Levison et al., Citation2016). E. coli, which has an isolation rate of 10.2% in bovine mastitis (Tenhagen, Hansen, Reinecke, Heuwieser, & Lam, Citation2009), can be classified into pathogenic and nonpathogenic bacteria, and pathogenic bacteria can be further classified into different types based on the pathogenic mechanism (Blum & Leitner, Citation2013)and virulence factor, such as enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), Shiga toxin-producing E. coli (STEC or VTEC), and enterohemorrhagic E. coli (EHEC (Fitzhenry et al., Citation2002; Nagy & Fekete, Citation1999)). STEC strains play important roles in mastitis (Güler & Gündüz, Citation2007; Kobori, Rigobelo, Macedo, Marin, & Avila, Citation2004) and EPEC strains are predominant in diarrheal patients, because different geographical location might influence the pathogenic molecular markers (Blanco, Blanco, Blanco, et al., Citation2004; Suardana, Artama, Asmara, & Dayono, Citation2010).
Bovine mastitis can be controlled and/or prevented by antimicrobial agents. Unfortunately, the misuse of antimicrobials has contributed to the emergence of antimicrobial-resistant bacteria (Mia et al., Citation2017; Van Boeckel et al., Citation2015; Zhang, Ying, Pan, Liu, & Zhao, Citation2015). For example, 20% to 33% of E. coli isolates from bovine mastitis were resistant to at least one antimicrobial agent (Fairbrother et al., Citation2015; Suojala et al., Citation2011), and 87.1% of E. coli isolates (61/70) are resistant to ampicillin and kanamycin according to a study performed in Beijing, China (Liu et al., Citation2014). Multidrug-resistant E. coli strains, which represent a significant public concern (Erb, Sturmer, Marre, & Brenner, Citation2007; Johnson et al., Citation2008), have been isolated from animals in several countries including China (Rao et al., Citation2014; Seni et al., Citation2016; Xu, An, Wang, & Zhang, Citation2015). Additionally, several virulence factors associated with E. coli infections and various pathogenic bacterial genes have been isolated from mastitis in dairy cows (Zhang et al., Citation2018). Virulence genes identified in E. coli from bovine mastitis include genes that code for hemagglutinin, aerobactin, P-fimbria, enterohemolysin, intimin, and autoagglutinating adhesion proteins (Bekal et al., Citation2003; Ewers et al., Citation2007; Johnson et al., Citation2008). The antimicrobial resistance and virulence genes of E. coli from bovine mastitis have been studied in Korea, Jordan, Iran, and Spain (Mora et al., Citation2005; Obaidat, Bani Salman, Davis, & Roess, Citation2018; Tark et al., Citation2017; Tavakoli & Pourtaghi, Citation2017; Zhao et al., Citation2018). A more thorough understanding of the resistance and virulence factors of E. coli isolates will assist in the implementation of appropriate treatments for the management of bovine mastitis.
Virulence factors may be associated with phylogeny groups (Garcia-Aljaro, Moreno, Andreu, Prats, & Blanch, Citation2009). Furthermore, the determination of phylogroups (Clermont et al., Citation2008), intimin typing (Blanco et al., Citation2005), and O serotyping (Iguchi et al., Citation2015) of E. coli can be used for pointed potential health risks. Researchers have screened virulence genes in E. coli isolated from poultry in Canada with O serotypes, unknown intimin types, and phylogroups (Afset et al., Citation2006). However, few studies have evaluated virulence factors and typing in mastitic cows. Therefore, the objectives of this study were to assess (i) antibiotic resistance of E. coli isolates, (ii) virulence gene profiles, (iii) phylogroups, (iv) O serogroups, and (v) intimin typing.
2. Material and methods
2.1. Sample collection
Five provinces, including Hebei, Shandong, Hei Longjiang, Shanghai, and Inner Mongolia were selected as the sampling points (2017), which covered four geographical locations in China. Four types of farms were selected based on the population of cows in each sampling point. Among the selected farms, a total of 497 raw milk samples were collected twice from cows with clinical mastitis in 2017 (May and September): 100 samples from Hebei province, 100 samples from Inner Mongolia, 100 samples from Shanghai city, 100 samples from Heilongjiang province, and 97 samples from Shandong province. We selected two specific months which represent summer and fall for collection of mastitis milks, because the average temperature of the five sampled provinces and cities we selected in May and September is different. Therefore, we plans to analyze whether there was an impact on E. coli resistance in bovine mastitis. All samples were transferred into sterile plastic bottles (Corning, New York, USA) and transported to the laboratory at 4°C within 4 h.
2.2. Isolation and identification of E. coli
The collected samples (25 mL) were homogenized in 225 mL of modified E. coli broth. Aliquots (1.0 mL) of selected dilutions were streak-inoculated in MacConkey agar plates (Huankai, Guangdong, China) and incubated for 18–24 h at 36 ± 1°C. Presumptive colonies were selected from the nutrient agar and placed in prepared sterile saline, resulting in a 0.5 McFarland standard. Aliquots (1.0 mL) of the bacterial suspension were pipetted into a micro-biochemical tube of the IMVC biochemical identification kit (Huankai, Guangdong, China) and incubated at 36°C. The detected E. coli was stored in a seed preservation tube containing 20%–30% glycerol in brain-heart extract broth and stored in the refrigerator at −80°C.
The molecular identification of isolated strains was conducted by PCR (Bio-Rad S1000, Bio-Rad Laboratories, Hercules, CA, USA) of 16s rDNA and rpoB (Dahllof, Baillie, & Kjelleberg, Citation2000). PCR samples were sequenced (HuaDa, Beijing, China) and compared using the BLAST programme at the National Center for Biotechnology Information website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The PCR reaction mixture (50 μL) consisted of 25 μL EmeraldAmp Max PCR Master Mix (Takara, Dalian, China), 19 μL dH2O (Takara, Dalian, China), 2 μL of each primer (100 μM; Sangon Biotech Shanghai Co., Ltd., Shanghai, China), and approximately 2 μL of bacterial genomic DNA (Supplemental Table S1). A negative control (without DNA template) and a positive control (E. coli ATCC 25922 template) were included in all PCR assays.
2.3. Antimicrobial susceptibility test
Identified E. coli isolates were tested for phenotypic susceptibility to antimicrobials using the MIC broth microdilution method (Romney, Citation2018). The drugs included ampicillin (4−16 μg/mL), amoxicillin-clavulanic acid (4/2–16/8 μg/mL), cefoxitin (1–16 μg/mL), tetracycline (2–8 μg/mL), chloramphenicol (4−16 μg/mL), gentamicin (2–8 μg/mL), trimethoprim/sulfamethoxazole (9.5–38 μg/mL), piperacillin (4–64 μg/mL), ciprofloxacin (0.5−2 μg/mL), aztreonam (2−16 μg/mL), meropenem (1–8 μg/mL), ofloxacin (1−8 μg/mL), and kanamycin (4–16 μg/mL). E. coli ATCC 25922 was used as a reference strain. Antimicrobial resistance was defined as non-susceptible to one or more antimicrobials, whereas multidrug resistance (MDR) was defined as non-susceptible to three and more classes of antimicrobials.
2.4. Detection of antimicrobial resistance genes
Phenotypically resistant E. coli isolates were characterized at the molecular level for their antimicrobial resistance mechanisms. PCR was performed as previously reported for the detection of drug-resistant genes associated with β-lactams (ampicillin, blaTEM , blaCTX-M, blaNDM , blaOXA and blaKPC), including cefoxitin (blaFOX). Amino acid changes in gyrA and gyrB proteins were studied by PCR and sequencing of the corresponding genes in quinolone-resistant isolates. Additionally, aminoglycoside (aac3, aacA4, and aac6-I), chloramphenicol (florR), and tetracycline (tetA and tetB) resistance genes were analyzed by PCR and sequenced. The amplified products were analyzed by electrophoresis in a 1.5% agarose gel stained with SYBR® Safe DNA gel stain (ThermoFisher Scientific, USA) and visualized using VersaDoc (Bio-Rad, Hercules, California, USA). The PCR reaction mixture (25 μL) consisted of 12.5 μL EmeraldAmp Max PCR Master Mix (Takara, Dalian, China), 9.5 μL dH2O (Takara, Dalian, China), 1.0 μL of each primer (100 μM; Sangon Biotech Shanghai Co., Ltd., Shanghai, China), and approximately 1.0 μL of bacterial genomic DNA (Supplemental Table S1).
2.4.1. PCR for virulence genes
Virulence genes including eae, aggR, esp, elt, ipaH, stx1, and stx2 were amplified by PCR (Bio-Rad). The primer sequences of all virulence genes used for PCR and the cycling conditions are shown in Supplementary Table S2. The PCR reaction mixture (25 μL) consisted of 12.5 μL EmeraldAmp Max PCR Master Mix (Takara, Dalian, China), 9.5 μL dH2O (Takara, Dalian, China), 1.0 μL of each primer (100 μM; Sangon Biotech Shanghai Co., Ltd., Shanghai, China), and approximately 1.0 μL of bacterial genomic DNA.
2.5. Typing methods
2.5.1. Phylogenetic grouping
The distributions of E. coli isolates in phylogroups were determined according to the phylogenetic grouping method for quadruplex PCR (Clermont et al., Citation2013a). The phylogenetic grouping method can assign E. coli strains into eight phylogroups (A, B1, B2, C, D, E, F, and clade-1). The data were used to adjust the primer sequences (chuA, yjaA, TspE4.C2, and arpA), and the sizes of PCR products were used for the quadruplex phylo-typing method. Analyses were performed using positive and negative controls. Primer sequences and cycling conditions for the phylo-typing method are shown in Supplemental Table S3.
2.5.2. Intimin typing
Complementary primers were used to identify 17 eae genes and intimin types by PCR. The primer sequences of intimin subtypes (α1, α2, β1, ξR/β2, δ/κ/β2O, γ1, θ/γ2, ε1, νR/ε2, ζ, η, ι1, μR/ι2, λ, μB, νB, and ξB) are shown in Supplemental Table S4 (Adu-Bobie et al., Citation1998; Blanco et al., Citation2004d; Blanco et al., Citation2005). The nucleotides sequence of the amplification products purified with a DNA purification kit (Qiagen) was determined by the deoxynucleotide triphosphate chain terminated method of Sanger using a BigDye Terminator v3.1 Cycle Sequencing kit and an ABI 3100 Genetic Analyzer (Applied Bio-Systems).
2.5.3. O-serogroups genes detection
The O-serotype genes of isolates were identified by PCR using seven pairs of primers to determine serogroups of E. coli (Supplemental Table S5).
2.6. Statistical analyses
Statistical analyses for each of the antimicrobial agents and resistance genes were performed. For phenotypic and genotypic analyses, data were analyzed by one-way ANOVA using SPSS (ver. 19.0, IBM Corp., Armonk, NY, USA). The results were presented as mean ± standard deviation. Paired t-tests were used for comparisons between groups. Statistical significance was set at P < 0.05.
3. Results
3.1. E. coli identification
There were 92 E. coli isolates in 497 raw milk samples. Among the isolates, 12 (13.0%) were from Shandong province, 18 (19.0%) were from Heilongjiang province, 19 (21.0%) were from Inner Mongolia, 19 (21.0%) were from Hebei province, and 24 (26.0%) were from Shanghai.
3.2. Antimicrobial susceptibility testing
The MIC broth microdilution method was used, and the interpretation of antibiotic breakpoint was carried out according to CLSI guidelines (Citation2018). A total of 13 drugs were tested in this study, and 48 of the 92 E. coli isolates were non-susceptible. The isolates were resistant to ampicillin (38/92, 41.3%), piperacillin (34/92, 36.9%), tetracycline (32/92, 34.8%), trimethoprim/sulfamethoxazole (31/92, 33.7%), chloramphenicol (23/92, 28.3%), aztreonam (18/92, 19.6%), gentamicin (17/92, 18.5%), ofloxacin (12/92, 13.0%), ciprofloxacin (8/92, 8.7%), cefoxitin (6/92, 6.5%), and amoxicillin-clavulanic acid (4/92, 4.3%). None of the isolates were resistant to meropenem or kanamycin (). The 48 isolates resistant to one or more antibiotics presented 37 different resistance patterns (). MDR was observed in 32/92 (34.8%) of the isolates, and two of the isolates were resistant to eight classes of antibiotics (). Furthermore, the level of antimicrobial resistance was higher in phylogroup B1 than in group A ().
Figure 1. Antibiotic resistance of E. coli isolates from bovine mastitis in five major dairy-producing regions of China.
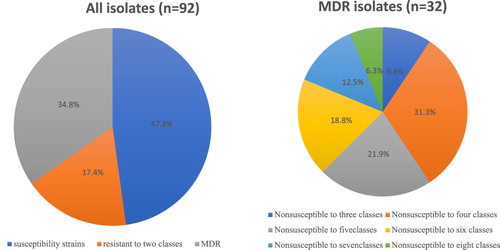
Table 1. Antibiotic resistance (%) of E. coli isolates from raw milk of mastitic cows in China.
Table 2. Antimicrobial resistance patterns of E. coli isolates.
Table 3. Percentage of resistant strains based on phylogenetic group.
3.3. Detection of antimicrobial resistance genes
In our study, 13 resistance genes associated with β-lactam, sulfonamide, tetracycline, and aminoglycoside antibiotics were analyzed (). E. coli isolates resistant to β-lactam including cefoxitin carried no blaKPC, blaFOX, blaNDM, or blaOXA genes. Among the isolates, 97.8% (90/92) carried gyrA, and 95.6% (84/92) carried gyrB. Approximately 75% (24/32) of tetracycline-resistant strains possessed tetA; however, no tetB was detected. Isolates with resistance to gentamicin contained aac4A (13/92, 14.1%), aac6 (9/92, 9.7%), and aac4A + aac6 (8/92, 8.7%), but none of the isolates contained aac3. Among the chloramphenicol-resistant isolates, 18 (78.0%) contained floR. Furthermore, 50.0% of penicillin-resistant strains had blaTEM, and 42.0% strains had blaCTX-M.
Table 4. Antimicrobial resistance genes identified in E. coli isolates from raw milk of mastitic cows.
3.4. Virulence distributions of E. coli isolates
The distribution of virulence genes in the 92 isolates is shown in . In our study, the eae gene was identified in 75 isolates, which were classified as EPEC, while no stx genes were detected. All eae gene-positive E. coli strains were divided into seven O-serogroups. The remaining 57 isolates possessed ipaH and were classified as EIEC, whereas six strains contained elt genes and were classified as ETEC.
Table 5. Prevalence of virulence genes in E. coli isolates.
3.5. Typing results
3.5.1. Phylogenetic typing
All strains were assigned to five groups according to Clermont’s phylogenetic method (Clermont et al., Citation2000). The distribution of serogroups, intimin types, and virulence genes among the different phylogenetic groups were determined. The majority of the E. coli strains were assigned to phylogroup B1 (35.9%), while the other strains were assigned to phylogroup A (31.5%), C (22.8%), or E (4.3%). However, five strains (5.4%) were not assigned to any phylogroup. Approximately 41.7% of Shandong strains were in phylogroup A, while group B1 strains were more predominant (45.8%) in Shanghai. Phylogroup E consisted of four strains from Hei Longjiang, Hebei, and Inner Mongolia ().
Table 6. Distribution of O serogroups among different phylogenetic groups.
3.5.2. O serotype grouping
A total of 65 of the 92 E. coli strains (70.7%) belonged to seven O serotypes (O121, O91, O22, O26, O128, O111, and O113), and the remaining 27 strains were untypeable because the serotypes could not be determined (UT; ). The prevalent serotypes were O121 and O91, accounting for 35.9% (33/92) and 16.3% (15/92), respectively. Other O genotypes obtained were O128 (seven strains, 7.6%), O22 (five strains, 5.4%), O111 and O113 (two strains in each genotype, 2.1%), and O26 (one strain, 1.1%). The isolates in phylogroups A and B1 had several O genotypes, whereas the C and E isolates were divided into four and two serogroups, respectively ().
3.5.3. Typing of intimin
Among the 92 E. coli strains, 75 (81.5%) were positive for the eae gene. Intimin-type eae-μR-ι2 was highly prevalent (48.9%,45/92) and belonged to six different O serogroups, additionally, 12 strains were designed UT, it was predominant (17/45, 15.6%) in phylogroup C compared other groups (A, B1, and E) (). Intimin type λ (16.3%, 15/92) belonged to four serogroups. Ten strains were detected in type ξR among E. coli strains subjected to three serogroups, three and two strains showed positive reactions using two groups of primers (νR-ε2 and η). The remaining E. coli strains were UT because amplicons were not generated using the typing primers ().
Figure 2. Distribution of intimin types among phylogroups in E. coli isolates from bovine mastitis in China.
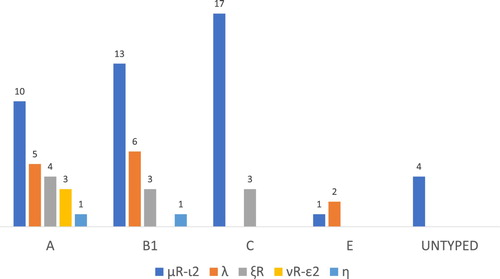
Table 7. Distribution of O serogroups among different intimin types.
4. Discussion
A recent report of large Chinese dairy farms indicated that E. coli is involved in the pathogenesis of bovine mastitis (Gao et al., Citation2017). In our study, there were 92 E. coli isolates (18.5%) in 497 raw milk samples. The prevalence of E. coli isolates was higher in our study than in a study performed in Turkey (3.2%, 8/250) (Akkaya, Cetinkaya, Alisarli, Telli, & Gök, Citation2008) and in Liaoning province, China (20.1%,79/389) (Zhang et al., Citation2018). Our results revealed that E. coli is prevalent in raw milk from mastitic cows in all five regions of China.
Antimicrobials are commonly used in the control and/or prevention of mastitis. Unfortunately, the misuse of antimicrobials contributes to the emergence of bacterial strains with antimicrobial resistance (Metzger & Hogan, Citation2013). In this study, a high percentage of strains were resistant to ampicillin (41.3%), followed by tetracycline (34.8%; ), possibly due to the overuse of drugs in the provinces we selected. The resistance rate towards ampicillin was higher in our study than in South Korea in 2012–2015 (16.6%) (Tark et al., Citation2017) and some provinces of China (25.3%) (Zhang et al., Citation2018). Antimicrobial resistance to ampicillin can be used to predict the susceptibility to amoxicillin (Romney, Citation2018). Therefore, it can be speculated that the E. coli strains showed a high resistance to amoxicillin. Resistance rates towards tetracycline were 9.0% in the European Union (Thomas et al., Citation2015), 14.3% in Canada (Saini et al., Citation2012), and 55.1% in Southwestern Nigeria (Adesoji, Ogunjobi, Olatoye, & Douglas, Citation2015). Moreover, 34.8% isolates had MDR (). A previous study reported that 45.8% of 48 E. coli strains were resistant to 13 antibiotics (Todorovic et al., Citation2018). The continuous use of antibiotics may cause MDR in dairy herds (Suojala et al., Citation2011). Therefore, it is important to control antibiotic use to prevent the risk of MDR. In Korea, researchers have concluded that the decline in tetracycline resistance may be due to the decreased use of the antibiotic in cattle (Tark et al., Citation2017).
Reports on antimicrobial resistance are useful for understanding the pathogenesis of E. coli infection in bovine mastitis (Blum et al., Citation2008). Broad-spectrum antibiotics, especially tetracyclines and quinolones, are frequently used in the clinical treatment of mastitis (Chajęcka-Wierzchowska, Zadernowska, & Łaniewska-Trokenheim, Citation2016; Zurfluh et al., Citation2014). It has been reported that resistance genotypes against tetracycline are correlated with resistance phenotypes in S. aureus and E. coli (Gomi et al., Citation2016; Yu et al., Citation2018). Some studies have shown that the tetA gene is more prevalent than the tetB gene in E. coli strains (Gomi et al., Citation2016; Hu et al., Citation2008; Rebbah et al., Citation2017), and our results supported this finding. In our study, 24 tetracycline-resistant isolates (75%) carried the tetracycline gene tetA; however, no tetB was detected. Our results revealed a discrepancy between genotype and phenotype. Quinolone resistance genes, gyrA and gyrB, were detected in more than 95% isolates, and only eight isolates had resistance to ciprofloxacin (), according to CLSI (Citation2018). The results revealed the differences between genotype and phenotype in ciprofloxacin-resistant E. coli strains. The misuse of antimicrobials is a contributing factor, but probably not the main or single factor involved in antimicrobial resistance (Miika, Nyberg, Pentti, Pirkko, & Hakanen, Citation2009; Oliver, Murinda, & Jayarao, Citation2011). Future studies should evaluate the differences between phenotype and genotype.
Another factor correlated with the pathogenesis of E. coli infections in bovine mastitis is virulence profile. In the present study, none of the E. coli isolates were STEC (). Similar findings were reported in Iran (Mansouri-Najand & Khalili, Citation2007) and Belgium (Vivegnis, Ellioui, Leclercq, & Lambert, Citation1999). However, low prevalence rates were found in milk from US (Jayarao et al., Citation2006; Van Kessel, Karns, Lombard, & Kopral, Citation2011) and Europe (Claeys et al., Citation2013; Little et al., Citation2008; Ombarak et al., Citation2016; Pradel, Bertin, Martin, & Livrelli, Citation2008). In this study, there were 75 EPEC strains, in agreement with the findings of Momtaz (Momtaz, Citation2010). However, EPEC prevalence in bovine mastitis was lower in Brazil (40.0%) than in our study (Schroeder et al., Citation2002). There might be a direct relationship between the presence of eae and the capacity of EPEC to cause disease.
Intimin type is an important E. coli virulence determinant because intimin is necessary in the pathogenesis of EPEC (Dean-Nystrom, Bosworth, Moon, & O’Brien, Citation1998). The intimin variation could cause specificity for different host tissues, and intimin type could be a key factor in animal-host specificity (Fitzhenry et al., Citation2002; Torres, Zhou, & Kaper, Citation2005; Zhang et al., Citation2002). Overall, the 75 EPEC strains had five different intimin types (μR-ι2, λ, ξR/β2, νR-ε2, and η). To the best of our knowledge, there have been no reports on intimin types in milk from mastitic cows (Blanco, Blanco, Dahbi, Mora, et al., Citation2006; Osman, Mustafa, Aly, & AbdElhamed, Citation2012). These five intimins have been isolated from fecal samples of adults and children with clinical diarrhea or other gastrointestinal alterations and from neotropical non-human primates with diarrhea (Blanco, Blanco, Blanco, et al., Citation2004; Blanco et al., Citation2005; Blanco, Blanco, Dahbi, Alonso, et al., Citation2006). Furthermore, intimin η was observed in bovine typical EPEC strains (Blanco et al., Citation2005; Blanco, Blanco, Dahbi, Alonso, et al., Citation2006). The findings reveal that different intimin EPEC strains are important pathogens associated with human diarrhea and bovine mastitis, which suggests that intimins may play a role in the transmission of infections between animals and humans.
Genotyping is a useful technique in epidemiology. In this study, we coupled serotyping with other molecular typing methods. Most of the seven O serogroups have not been detected in bovine cows. Approximately 77.4% of E. coli isolates belonged to four different O serogroups (O26, O86, O111, and O127) in Egypt (Osman et al., Citation2012). The high frequency of eae found in this study may be related to the high frequency of certain serogroups, as it has been reported that the presence of eae is associated with specific O serogroups, such as O157, O145, O103, O26, and O111 (Miika et al., Citation2009). In this study, 45 intimin μR-ι2 isolates were in six serotypes (O121, O91, O113, O128, O22, and O111), 15 intimin λ strains were in four serotypes (O121, O91, O22, and O128), 10 intimin ξR/β2 strains were in three serotypes (O26, O91, O121), and intimin η was in serotype O121. O91 and O121 were frequently observed in these five intimin types. Our results showed that serogroups, such as O26, O91, O111, O113, O121, and O145 may cause diarrhea (Kappeli, Hachler, Giezendanner, Beutin, & Stephan, Citation2011; Schmidt, Beutin, & Karch, Citation1995). Furthermore, intimin types (β1 and γ1) present in O157 and O26 were similar in human and bovine tissue (Blanco, Blanco, Mora, et al., Citation2004). Therefore, isolates from milk, which belong to serogroups associated with diarrhea, are potentially pathogenic to humans.
In this study, E. coli strains associated with mastitis belonged to groups B1 (35.9%) and A (31.5%). In Brazil, most mastitis E. coli isolates were assigned to phylogroups A (52%) and B1 (38%) (Tomazi, Coura, Goncalves, Heinemann, & Santos, Citation2018). The major antibiotic-resistant isolates belonged to phylogroups A and B1, and phylogroup B1 had a higher level of antimicrobial resistance (). Additionally, 32 isolates of the EPEC strains were resistant to one or more antibiotics, and 23 isolates were non-susceptible to three or more antibiotic classes (Supplemental Figure 1). In Beijing, 58.6% of antibiotic-resistant E. coli strains were in group B1 and 35.7% were in group A (Garcia-Aljaro et al., Citation2009). Additionally, E. coli isolates with MDR were classified in phylogenetic groups A or B1 (Ramos et al., Citation2013). The combination of different virulence determinants and the phylogeny of bacteria may improve the recognition of new subgroups of virulent bacteria (Picard et al., Citation1999). The 75 EPEC strains in our study harbouring intimin genes could be separated into groups A (30.7%, 23/75), B1 (30.7%, 23/75), C (26.7%, 20/75), and E (4.0%, 3/75) (). Similarly, researchers reported that intimin type β1 in the majority of E. coli from poultry fecal samples belonged to phylogroup B1 and virulence group I (Parvej et al., Citation2019). The intimin μR-ι2 isolates in this study were most frequently found in phylogroup C.
5. Conclusions
The antimicrobial resistance of E. coli strains in the five major dairy-producing regions in China should be a public concern. E. coli strains were resistant against antibiotics especially ampicillin, but susceptible to meropenem and kanamycin. Future studies should evaluate the relationship among antimicrobial susceptibility, phylogroups, and virulence genes of E. coli strains associated with human and bovine mastitis. Additionally, studies should assess whether EPEC isolated from bovine mastitis may cause disease in humans.
Ethics approval and consent to participate
All experimental procedures involving the care and management of dairy cows, and the collection of bovine mastitis milk in the current study, were approved by the Animal Care and Welfare Committee of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences.
Supplemental Material
Download MS Word (28.7 KB)Acknowledgement
We sincerely thank all the participants who took part in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adesoji, A. T., Ogunjobi, A. A., Olatoye, I. O., & Douglas, D. R. (2015). Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria. Annals of Clinical Microbiology & Antimicrobials, 14, 35–42. doi: 10.1186/s12941-015-0093-1
- Adu-Bobie, J., Frankel, G., Bain, C., Goncalves, A. G., Trabulsi, L. R., Douce, G. et al. (1998). Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. Journal of Clinical Microbiology 36, 662–668. doi: 10.1128/JCM.36.3.662-668.1998
- Afset, J. E., Bruant, G., Brousseau, R., Harel, J., Anderssen, E., Bevanger, L., & Bergh, K. (2006). Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. Journal of Clinical Microbiology, 44, 3703–3711. doi: 10.1128/JCM.00429-06
- Akkaya, L., Cetinkaya, Z., Alisarli, M., Telli, R., & Gök, V. (2008). The prevalence of E. coli O157/O157: H7, L. monocytogenes and Salmonella spp. on bovine carcasses in Turkey. Journal of Muscle Foods, 19, 420–429. doi: 10.1111/j.1745-4573.2008.00127.x
- Bekal, S., Brousseau, R., Masson, L., Prefontaine, G., Fairbrother, J., & Harel, J. (2003). Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. Journal of Clinical Microbiology, 41, 2113–2125. doi: 10.1128/JCM.41.5.2113-2125.2003
- Blanco, M., Blanco, J. E., Blanco, J., de Carvalho, V. M., Onuma, D. L., & Pestana de Castro, A. F. (2004). Typing of intimin (eae) genes in attaching and effacing Escherichia coli strains from monkeys. Journal of Clinical Microbiology, 42, 1382–1383. doi: 10.1128/JCM.42.3.1382-1383.2004
- Blanco, M., Blanco, J. E., Dahbi, G., Alonso, M. P., Mora, A., Coira, M. A., … Blanco, J. (2006). Identification of two new intimin types in atypical enteropathogenic Escherichia coli. International Microbiology, 9, 103–110.
- Blanco, M., Blanco, J. E., Dahbi, G., Mora, A., Alonso, M. P., Varela, G., … Blanco, J. (2006). Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: Identification of two novel intimin variants (muB and xiR/beta2B). Journal of Medical Microbiology, 55, 1165–1174. doi: 10.1099/jmm.0.46518-0
- Blanco, M., Blanco, J. E., Mora, A., Dahbi, G., Alonso, M. P., González, E. A., … Blanco, J. (2004). Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). Journal of Clinical Microbiology, 42, 645–651. doi: 10.1128/JCM.42.2.645-651.2004
- Blanco, M., Padola, N. L., Kruger, A., Sanz, M. E., Blanco, J. E., Gonzalez, E. A. et al. (2004d). Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int Microbiol 7, 269–276.
- Blanco, M., Schumacher, S., Tasara, T., Zweifel, C., Blanco, J. E., Dahbi, G., … Stephan, R. (2005). Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-η2). BMC Microbiology, 5, 23–33. doi: 10.1186/1471-2180-5-23
- Blum, S., Heller, E. D., Krifucks, O., Sela, S., Hammer-Muntz, O., & Leitner, G. (2008). Identification of a bovine mastitis Escherichia coli subset. Veterinary Microbiology, 132, 135–148. doi: 10.1016/j.vetmic.2008.05.012
- Blum, S. E., & Leitner, G. (2013). Genotyping and virulence factors assessment of bovine mastitis Escherichia coli. Veterinary Microbiology, 163, 305–312. doi: 10.1016/j.vetmic.2012.12.037
- Chajęcka-Wierzchowska, W., Zadernowska, A., & Łaniewska-Trokenheim, Ł. (2016). Virulence factors, antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from retail shrimps. LWT - Food Science and Technology, 69, 117–122. doi: 10.1016/j.lwt.2016.01.034
- Claeys, W. L., Cardoen, S., Daube, G., De Block, J., Dewettinck, K., Dierick, K., … Herman, L. (2013). Raw or heated cow milk consumption: Review of risks and benefits. Food Control, 31, 251–262. doi: 10.1016/j.foodcont.2012.09.035
- Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013a). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports 5, 58–65. doi: 10.1111/1758-2229.12019
- Clermont, O., Lescat, M., O’Brien, C. L., Gordon, D. M., Tenaillon, O., & Denamur, E. (2008). Evidence for a human-specific Escherichia coli clone. Environmental Microbiology, 10, 1000–1006. doi: 10.1111/j.1462-2920.2007.01520.x
- Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
- Dahllof, I., Baillie, H., & Kjelleberg, S. (2000). rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Applied and Environmental Microbiology, 66, 3376–3380. doi: 10.1128/AEM.66.8.3376-3380.2000
- Dean-Nystrom, E. A., Bosworth, B. T., Moon, H. W., & O’Brien, A. D. (1998). Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infection and Immunity, 66, 4560–4563. doi: 10.1128/IAI.66.9.4560-4563.1998
- Erb, A., Sturmer, T., Marre, R., & Brenner, H. (2007). Prevalence of antibiotic resistance in Escherichia coli: Overview of geographical, temporal, and methodological variations. European Journal of Clinical Microbiology & Infectious Diseases, 26, 83–90. doi: 10.1007/s10096-006-0248-2
- Ewers, C., Li, G., Wilking, H., Kiessling, S., Alt, K., Antao, E., … Homeier, T. (2007). Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? International Journal of Medical Microbiology, 297, 163–176. doi: 10.1016/j.ijmm.2007.01.003
- Fairbrother, J. H., Dufour, S., Fairbrother, J. M., Francoz, D., Nadeau, E., & Messier, S. (2015). Characterization of persistent and transient Escherichia coli isolates recovered from clinical mastitis episodes in dairy cows. Veterinary Microbiology, 176, 126–133. doi: 10.1016/j.vetmic.2014.12.025
- Fitzhenry, R. J., Pickard, D. J., Hartland, E. L., Reece, S., Dougan, G., Phillips, A. D., & Frankel, G. (2002). Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut, 50, 180–185. doi: 10.1136/gut.50.2.180
- Gao, J., Barkema, H. W., Zhang, L., Liu, G., Deng, Z., Cai, L., … Han, B. (2017). Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. Journal of Dairy Science, 100, 4797–4806. doi: 10.3168/jds.2016-12334
- Garcia-Aljaro, C., Moreno, E., Andreu, A., Prats, G., & Blanch, A. R. (2009). Phylogroups, virulence determinants and antimicrobial resistance in stx(2) gene-carrying Escherichia coli isolated from aquatic environments. Research in Microbiology, 160, 585–591. doi: 10.1016/j.resmic.2009.08.004
- Gomi, R., Matsuda, T., Matsumura, Y., Yamamoto, M., Tanaka, M., Ichiyama, S., & Yoneda, M. (2016). Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in River Water. Applied and Environmental Microbiology, 83(5), 3–16.
- Güler, L., & Gündüz, K. (2007). Virulence properties of Escherichia coli isolated from clinical bovine mastitis. Turkish Journal of Veterinary and Animal Sciences, 31, 361–365.
- Halasa, T., Huijps, K., Østerås, O., & Hogeveen, H. (2007). Economic effects of bovine mastitis and mastitis management: A review. Veterinary Quarterly, 29, 18–31. doi: 10.1080/01652176.2007.9695224
- Hu, J., Shi, J., Chang, H., Li, D., Yang, M., & Kamagata, Y. (2008). Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environmental Science & Technology, 42, 3415–3420. doi: 10.1021/es7026746
- Iguchi, A., Iyoda, S., Seto, K., Morita-Ishihara, T., Scheutz, F., Ohnishi, M., & Pathogenic E. coli Working Group in Japan. (2015). Escherichia coli O-Genotyping PCR: A comprehensive and practical platform for molecular O serogrouping. Journal of Clinical Microbiology, 53, 2427–2432. doi: 10.1128/JCM.00321-15
- Jayarao, B. M., Donaldson, S. C., Straley, B. A., Sawant, A. A., Hegde, N. V., & Brown, J. L. (2006). A survey of foodborne pathogens in bulk tank milk and raw milk consumption among farm families in pennsylvania. Journal of Dairy Science, 89, 2451–2458. doi: 10.3168/jds.S0022-0302(06)72318-9
- Johnson, T. J., Wannemuehler, Y., Johnson, S. J., Stell, A. L., Doetkott, C., Johnson, J. R., … Nolan, L. K. (2008). Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Applied and Environmental Microbiology, 74, 7043–7050. doi: 10.1128/AEM.01395-08
- Kappeli, U., Hachler, H., Giezendanner, N., Beutin, L., & Stephan, R. (2011). Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerging Infectious Diseases, 17, 180–185. doi: 10.3201/eid1702.100909
- Kobori, D., Rigobelo, E. C., Macedo, C., Marin, J. M., & Avila, F. A. (2004). Virulence properties of Shiga toxin-producing Escherichia coli isolated from cases of bovine Mastitis in Brazil = Propiedades de virulencia de la Escherichia coli productora de toxina Shiga aislada de vacas de leche con mastitis en Brasil = Caractéristiques de la virulence d’Escherichia coli, productrice de la toxine type Shiga, isolée à partir de cas de mammite bovine au Brésil. Revue d’Elevage et de Medecine Veterinaire des pays Tropicaux, 57(1-2), 15–20. doi: 10.19182/remvt.9899
- Levison, L. J., Miller-Cushon, E. K., Tucker, A. L., Bergeron, R., Leslie, K. E., Barkema, H. W., & DeVries, T. J. (2016). Incidence rate of pathogen-specific clinical mastitis on conventional and organic Canadian dairy farms. Journal of Dairy Science, 99, 1341–1350. doi: 10.3168/jds.2015-9809
- Little, C. L., Rhoades, J. R., Sagoo, S. K., Harris, J., Greenwood, M., Mithani, V., & McLauchlin, J. (2008). Microbiological quality of retail cheeses made from raw, thermized or pasteurized milk in the UK. Food Microbiology, 25, 304–312. doi: 10.1016/j.fm.2007.10.007
- Liu, Y., Liu, G., Liu, W., Liu, Y., Ali, T., Chen, W., … Han, B. (2014). Phylogenetic group, virulence factors and antimicrobial resistance of Escherichia coli associated with bovine mastitis. Research in Microbiology, 165, 273–277. doi: 10.1016/j.resmic.2014.03.007
- Mansouri-Najand, L., & Khalili, M. (2007). Detection of shiga-like toxigenic Escherichia coli from raw milk cheeses produced in Kerman-Iran. Veterinarski Arhiv, 77, 515–522.
- Metzger, S. A., & Hogan, J. S. (2013). Short communication: Antimicrobial susceptibility and frequency of resistance genes in Escherichia coli isolated from bovine mastitis. Journal of Dairy Science, 96, 3044–3049. doi: 10.3168/jds.2012-6402
- Mia, M. T., Hossain, M. K., Rumi, N. A., Rahman, M. S., Mahmud, M. S., & Das, M. (2017). Detection of bacterial species from clinical mastitis in dairy cows at Nilphamari district and their antibiogram studies. Asian Journal of Medical and Biological Research, 2, 656–663. doi: 10.3329/ajmbr.v2i4.31011
- Miika, B., Nyberg, S. T., Pentti, H., Pirkko, P., & Hakanen, A. J. (2009). Association between antimicrobial consumption and resistance in Escherichia coli. Antimicrobial Agents and Chemotherapy, 53, 912–917. doi: 10.1128/AAC.00856-08
- Momtaz, H. (2010). Investigation of virulence factors in escherichia coli isolated from clinical and subclinical bovine mastitis. Bulgarian Journal of Veterinary Medicine, 13(2), 122–126.
- Mora, A., Blanco, J. E., Blanco, M., Alonso, M. P., Dhabi, G., Echeita, A., … Blanco, J. (2005). Antimicrobial resistance of Shiga toxin (verotoxin)-producing Escherichia coli O157:H7 and non-O157 strains isolated from humans, cattle, sheep and food in Spain. Research in Microbiology, 156, 793–806. doi: 10.1016/j.resmic.2005.03.006
- Nagy, B., & Fekete, P. Z. (1999). Enterotoxigenic Escherichia coli (ETEC) in farm animals. Veterinary Research, 30, 259–284.
- Obaidat, M. M., Bani Salman, A. E., Davis, M. A., & Roess, A. A. (2018). Major diseases, extensive misuse, and high antimicrobial resistance of Escherichia coli in large- and small-scale dairy cattle farms in Jordan. Journal of Dairy Science, 101, 2324–2334. doi: 10.3168/jds.2017-13665
- Olde Riekerink, R. G., Barkema, H. W., Kelton, D. F., & Scholl, D. T. (2008). Incidence rate of clinical mastitis on Canadian dairy farms. Journal of Dairy Science, 91, 1366–1377. doi: 10.3168/jds.2007-0757
- Olde Riekerink, R. G., Barkema, H. W., Scholl, D. T., Poole, D. E., & Kelton, D. F. (2010). Management practices associated with the bulk-milk prevalence of Staphylococcus aureus in Canadian dairy farms. Preventive Veterinary Medicine, 97, 20–28. doi: 10.1016/j.prevetmed.2010.07.002
- Oliver, S. P., Murinda, S. E., & Jayarao, B. M. (2011). Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: A comprehensive review. Foodborne Pathogens and Disease, 8, 337–355. doi: 10.1089/fpd.2010.0730
- Ombarak, R. A., Hinenoya, A., Awasthi, S. P., Iguchi, A., Shima, A., Elbagory, A. R., & Yamasaki, S. (2016). Prevalence and pathogenic potential of Escherichia coli isolates from raw milk and raw milk cheese in Egypt. International Journal of Food Microbiology, 221, 69–76. doi: 10.1016/j.ijfoodmicro.2016.01.009
- Osman, K. M., Mustafa, A. M., Aly, M. A., & AbdElhamed, G. S. (2012). Serotypes, virulence genes, and intimin types of shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli isolated from mastitic milk relevant to human health in Egypt. Vector-Borne and Zoonotic Diseases, 12, 297–305. doi: 10.1089/vbz.2010.0257
- Parvej, M. S., Nakamura, H., Alam, M. A., Wang, L., Zhang, S., Emura, K., … Dudley, E. G. (2019). Host range-associated clustering based on multilocus variable-number tandem-repeat analysis, phylotypes, and virulence genes of atypical enteropathogenic Escherichia coli strains. Applied and Environmental Microbiology, 85(6), 18–30. doi: 10.1128/AEM.02796-18
- Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., … Denamur, E. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and Immunity, 67, 546–553. doi: 10.1128/IAI.67.2.546-553.1999
- Pradel, N., Bertin, Y., Martin, C., & Livrelli, V. (2008). Molecular analysis of shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Applied and Environmental Microbiology, 74, 2118–2128. doi: 10.1128/AEM.02688-07
- Ramos, S., Silva, N., Canica, M., Capelo-Martinez, J. L., Brito, F., Igrejas, G., & Poeta, P. (2013). High prevalence of antimicrobial-resistant Escherichia coli from animals at slaughter: A food safety risk. Journal of the Science of Food and Agriculture, 93, 517–526. doi: 10.1002/jsfa.5814
- Rao, L., Lv, L., Zeng, Z., Chen, S., He, D., Chen, X., … Liu, J.-H. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Veterinary Microbiology, 172, 534–541. doi: 10.1016/j.vetmic.2014.06.013
- Rebbah, N., Messai, Y., Chatre, P., Haenni, M., Madec, J. Y., & Bakour, R. (2017). Diversity of CTX-M extended-spectrum beta-lactamases in Escherichia coli isolates from retail raw ground beef: First report of CTX-M-24 and CTX-M-32 in Algeria. Microbial Drug Resistance, 24, 896–908. doi: 10.1089/mdr.2017.0171
- Romney M. Humphries, J. A., Stephanie, L. M., Castanheira, M., Dingle, T., Hindler, J. A., Koeth, L., Sei, K., and on behalf of the CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. (2018). CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. Journal of Clinical Microbiology 56 : 1934–17.
- Saini, V., Mcclure, J. T., Léger, D., Keefe, G. P., Scholl, D. T., Morck, D. W., & Barkema, H. W. (2012). Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. Journal of Dairy Science, 95, 4319–4332. doi: 10.3168/jds.2012-5373
- Schmidt, H., Beutin, L., & Karch, H. (1995). Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infection and Immunity, 63, 1055–1061. doi: 10.1128/IAI.63.3.1055-1061.1995
- Schroeder, C. M., Zhao, C., Debroy, C., Torcolini, J., Zhao, S., White, D. G., … Meng, J. (2002). Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Applied & Environmental Microbiology, 68, 576–581. doi: 10.1128/AEM.68.2.576-581.2002
- Seni, J., Falgenhauer, L., Simeo, N., Mirambo, M. M., Imirzalioglu, C., Matee, M., … Mshana, S. E. (2016). Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Frontiers in Microbiology, 7, 142–149. doi: 10.3389/fmicb.2016.00142
- Suardana, I. W., Artama, W. T., Asmara, W., & Dayono, B. S. (2010). Adherence pheno-genotypic of Escherichia coli O157: H7 isolated from beef, feces of cattle, chicken and human. Indonesian Journal of Biotechnology, 16, 46–52.
- Suojala, L., Pohjanvirta, T., Simojoki, H., Myllyniemi, A. L., Pitkala, A., Pelkonen, S., & Pyorala, S. (2011). Phylogeny, virulence factors and antimicrobial susceptibility of Escherichia coli isolated in clinical bovine mastitis. Veterinary Microbiology, 147, 383–388. doi: 10.1016/j.vetmic.2010.07.011
- Tark, D. S., Moon, D. C., Kang, H. Y., Kim, S. R., Nam, H. M., Lee, H. S., … Lim, S.-K. (2017). Antimicrobial susceptibility and characterization of extended-spectrum beta-lactamases in Escherichia coli isolated from bovine mastitic milk in South Korea from 2012 to 2015. Journal of Dairy Science, 100, 3463–3469. doi: 10.3168/jds.2016-12276
- Tavakoli, M., & Pourtaghi, H. (2017). Molecular detection of virulence genes and multi-drug resistance patterns in Escherichia coli (STEC) in clinical bovine mastitis: Alborz province, Iran. Iranian Journal of Veterinary Research, 18, 208–211.
- Tenhagen, B. A., Hansen, I., Reinecke, A., Heuwieser, W., & Lam, T. J. G. M. (2009). Prevalence of pathogens in milk samples of dairy cows with clinical mastitis and in heifers at first parturition. Journal of Dairy Research, 76, 179–187. doi: 10.1017/S0022029908003786
- Thomas, V., de Jong, A., Moyaert, H., Simjee, S., El Garch, F., Morrissey, I., … Vallé, M. (2015). Antimicrobial susceptibility monitoring of mastitis pathogens isolated from acute cases of clinical mastitis in dairy cows across Europe: VetPath results. International Journal of Antimicrobial Agents, 46, 13–20. doi: 10.1016/j.ijantimicag.2015.03.013
- Todorovic, D., Velhner, M., Grego, E., Vidanovic, D., Milanov, D., Krnjaic, D., & Kehrenberg, C. (2018). Molecular characterization of multidrug-resistant escherichia coli isolates from bovine clinical mastitis and pigs in the Vojvodina Province, Serbia. Microbial Drug Resistance, 24, 95–103. doi: 10.1089/mdr.2017.0016
- Tomazi, T., Coura, F. M., Goncalves, J. L., Heinemann, M. B., & Santos, M. V. (2018). Antimicrobial susceptibility patterns of Escherichia coli phylogenetic groups isolated from bovine clinical mastitis. Journal of Dairy Science, 101, 9406–9418. doi: 10.3168/jds.2018-14485
- Torres, A. G., Zhou, X., & Kaper, J. B. (2005). Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infection and Immunity, 73, 18–29. doi: 10.1128/IAI.73.1.18-29.2005
- Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., … Laxminarayan, R. (2015). Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences, 112, 5649–5654. doi: 10.1073/pnas.1503141112
- Van Kessel, J. A., Karns, J. S., Lombard, J. E., & Kopral, C. A. (2011). Prevalence of Salmonella enterica, Listeria monocytogenes, and Escherichia coli virulence factors in bulk tank milk and in-line filters from U.S. dairies. Journal of Food Protection, 74, 759–768. doi: 10.4315/0362-028X.JFP-10-423
- Vivegnis, J., Ellioui, M., Leclercq, A., & Lambert, B. (1999). Detection of Shiga-like toxin producing Escherichia coli from raw milk cheeses produced in Wallonia [Belgium]. Biotechnologie Agronomie Société Et Environnement, 3, 7695–7724.
- Xu, G., An, W., Wang, H., & Zhang, X. (2015). Prevalence and characteristics of extended-spectrum β-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Frontiers in Microbiology, 6, 1103–1111.
- Yu, L., Li, W., Zhang, M., Cui, Y., Chen, X., Ni, J., … Xue, T. (2018). Imidazole decreases the ampicillin resistance of an Escherichia coli strain isolated from a cow with mastitis by inhibiting the function of autoinducer 2. Journal of Dairy Science, 101, 3356–3362. doi: 10.3168/jds.2017-13761
- Zhang, W. L., Köhler, B., Oswald, E., Beutin, L., Karch, H., Morabito, S., … Schmidt, H. (2002). Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. Journal of Clinical Microbiology, 40, 4486–4492. doi: 10.1128/JCM.40.12.4486-4492.2002
- Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., & Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environmental Science & Technology, 49, 6772–6782. doi: 10.1021/acs.est.5b00729
- Zhang, D., Zhang, Z., Huang, C., Gao, X., Wang, Z., Liu, Y., … Liu, M. (2018). The phylogenetic group, antimicrobial susceptibility, and virulence genes of Escherichia coli from clinical bovine mastitis. Journal of Dairy Science, 101, 572–580. doi: 10.3168/jds.2017-13159
- Zhao, Q. Y., Yuan, F. W., Liang, T., Liang, X. C., Luo, Y. R., Jiang, M., … Zhang, W. M. (2018). Baicalin inhibits Escherichia coli isolates in bovine mastitic milk and reduces antimicrobial resistance. Journal of Dairy Science, 101, 2415–2422. doi: 10.3168/jds.2017-13349
- Zurfluh, K., Abgottspon, H., Hächler, H., Nüeschinderbinen, M., Stephan, R., & Khan, A. U. (2014). Quinolone resistance mechanisms among extended-spectrum beta-lactamase (ESBL) producing Escherichia coli isolated from rivers and lakes in Switzerland. Plos One, 9, e95864. doi: 10.1371/journal.pone.0095864
