Abstract
Species level taxonomy and phylogeographical distribution patterns in the freshwater rhodophyte Sirodotia are resolved through phylogenetic inferences based on rbcL and cox2–3 sequence data. Previous studies focused on the taxonomy of specific Sirodotia species or the distributions across a limited geographical region. Our molecular phylogenies included samples attributable to five recognized Sirodotia species and include collections from Australia, Brazil, Costa Rica, Canada, Finland, Mexico, New Zealand, South Africa and the United States. Both rbcL and cox2–3 phylogenies inferred S. suecica, S. tenuissima and S. goebelii as a monophyletic group with little sequence divergence. This result supports the synonymy of S. tenuissima and S. goebelii with S. suecica (the species name with priority). Within this clade, samples collected from Australia and New Zealand formed a monophyletic group with no other discernible phylogeographical patterns within S. suecica. This result seems to be somewhat unusual in the Batrachospermales, as other species have shown greater genetic variation among geographically distant locations. As in previous studies, S. huillensis and S. delicatula were inferred as a separate species based on the rbcL phylogeny, supporting the current taxonomy. A specimen of S. aff. huillensis from South Africa, may represent a new species but further research is necessary before it can be designated as such.
Introduction
The red algal genus Sirodotia is a member of the strictly freshwater order Batrachospermales, similar in gross morphology to the genera Batrachospermum and Kumanoa. However, it is distinguished from these genera and all others in the order by two reproductive characteristics: an asymmetrical carpogonium base, and indeterminate gonimoblast filaments in the carposporophyte (Starmach, Citation1977; Necchi et al., Citation1993; Kumano, Citation2002). Species have been delineated based on a number of characters including gross morphology of the gametophyte, spermatangia on specialized branches, the position of the carpogonial bearing branch, which side of the carpogonium the gonimoblast filaments arise, and the size of the carpogonium (Starmach, Citation1977; Necchi et al., Citation1993, Citation2007). Although there are 13 infrageneric taxon names in use within the genus Sirodotia (Guiry & Guiry, Citation2011), Kumano (Citation2002) in his review of the freshwater red algae of the world recognized eight taxa, of which five were known only from the type locality and one or two other locations. Only S. suecica, S. huillensis and S. delicatula are reported as widespread.
The genus Sirodotia is found on all continents except Antarctica, having been collected from numerous locations in North America, South America, Australasia and northern Europe (including a single location in the British Isles), with scattered reports in China, India, south-east Asia and Africa (West & West, Citation1897; Israelson, Citation1942; Balakrishnan & Chaugule, Citation1980; Kumano, Citation1982; Necchi et al., Citation1993; Entwisle & Foard, Citation1999; Sheath & Sherwood, Citation2002; Carmona et al., Citation2006; Eloranta & Kwandrans, Citation2007; Necchi et al., Citation2007; Eloranta et al., Citation2011). Sirodotia suecica appears to be most abundant in northern temperate or boreal areas of North America and northern Europe, but also has been collected throughout eastern Australia and New Zealand in the southern hemisphere (Israelson, Citation1942; Necchi et al., Citation1993; Entwisle & Foard, Citation1999; Eloranta & Kwandrans, Citation2007). Sirodotia delicatula has been reported from Brazil in South America, Costa Rica in North America, Japan, Indonesia and Malaysia in Southeast Asia (Umezaki, Citation1960; Kumano, Citation1982; Necchi et al., Citation1999, Citation2007). The reported distribution for S. huillensis is the south-western USA (Arizona and Texas) in North America, China, Central Mexico, India and the type locality in Africa (West & West, Citation1897; Balakrishnan & Chaugule, Citation1980 [as ‘S. huiellense’]; Sheath et al., Citation1996; Vis et al., Citation1998; Kumano, Citation2002; Carmona et al., Citation2006; Hu & Wei, Citation2006). All three taxa seem to have wide and partially overlapping ranges.
Previous molecular studies of Sirodotia have provided insights into the infrageneric taxonomy. Vis & Sheath (Citation1999) analysed S. suecica, S. tenuissima and S. huillensis from a few sites in North America and determined S. tenuissima to be synonymous with S. suecica, but S. huillensis to be a distinct species. A study of the Batrachospermales in Australia revealed a single sample of S. suecica from that continent showed little sequence divergence from North American specimens, potentially suggesting it may be more readily dispersed than previously thought (Vis & Entwisle, Citation2000). More recently, sequence data for S. delicatula was added to the phylogeny, showing this species to be distinct, and to include a specimen from Costa Rica misidentified as S. huillensis (Necchi et al., Citation2007).
Some taxonomic changes have not been accepted by subsequent researchers. For example, S. tenuissima is still recognized by some authors due to its distinct morphology (Kwandrans & Eloranta, Citation2002; Eloranta & Kwandrans, Citation2007; Eloranta et al., Citation2011). Given that its synonymy with S. suecica was based on a single specimen from North America (Vis & Sheath, Citation1999), this decision would in any case benefit from further corroboration, using more specimens from a broader geographical area.
All previous molecular research has used a limited number of specimens from North America, South America and Australia, and the opportunity arose to include collections from two continents not previously represented, Europe and Africa, as well as new distant locations from North America and Australia. In addition, a specimen of S. goebelii from what is surmised to be the type locality (see Appendix: Supplementaryfile) was analysed to determine its phylogenetic affinity. The hypothesis that S. tenuissima and S. suecica are conspecific was tested with more specimens fitting the morphological description of both taxa and another well-studied intraspecific marker.
Materials and methods
Thirteen newly collected specimens of Sirodotia were obtained from Finland (6), Australia (1), New Zealand (1), South Africa (2) and USA (3) (). These specimens were identified using the morphological characteristics provided in protologues and other literature (Kylin, Citation1912; Skuja, Citation1938; Flint, Citation1948; Starmach, Citation1977; Necchi et al., Citation1993; Entwisle & Foard, Citation1999; Kumano, Citation2002). Only one specimen could not be positively identified due to the lack of defining characters and this was assigned the epithet S. aff. huillensis.
Table 1. Collection information, Genbank accession numbers and previous studies for specimens of Sirodotia used in this study. New sequences in bold.
A portion of each specimen was cleaned of visible epiphytes and preserved in silica desiccant for DNA extraction. Specimens were ground in liquid nitrogen and extracted using either the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) or NucleoSpin® (Clontech, Mountain View, CA, USA) following the manufacturer's protocols. For phylogenetic analyses, the plastid encoded rbcL gene was PCR amplified using the F160 and rbcLR primers (Vis et al., Citation1998) in a PCR cocktail that also included AmpliTaq Gold® Fast PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and Sirodotia DNA. Amplified PCR products were sized on 1.6% Tris-Borate-EDTA (TBE) agarose gels. Products were purified for DNA sequencing using the Ultraclean PCR Clean-Up Kit (Mo Bio Laboratories, Carlsbad, CA, USA) following the manufacturer's protocols. The forward and reverse PCR primers were added to separate DNA sequencing reactions. In order to sequence the 1282 bp rbcL fragment completely in both directions, the following internal forward (F) and reverse (R) primers were used for DNA sequencing: F1087.wbc1 (5′–GTCATCTAGATGTTAATTTACCTC–3′), R472.4, R897 (Vis & Sheath, Citation1999), F650, R897.1 (Vis et al., Citation2010). Internal primer selection was based on examination of F160 sequence chromatographs. To study the phylogeographical distribution pattern of S. suecica, the mitochondrial cox2–3 spacer region was amplified using the cox2F and cox3R primers (Zuccarello et al., Citation1999). PCR conditions for each marker followed Vis & Entwisle (Citation2000). PCR products were prepared and sequenced as in Vis et al. (Citation2010). Sequence data were assembled in Sequencher™4.10.1 (Gene Codes, Ann Arbor, MI, USA). Newly generated sequence data from this study were submitted to GenBank ().
Phylogenetic analyses were conducted separately on the rbcL and cox2–3 datasets. Maximum likelihood (ML) and Bayesian inference (BI) trees were constructed for each dataset. Nucleotide substitution models for both the ML and BI analyses were selected by jModeltest (Posada, Citation2008) via the corrected Akaike Information Criterion (AICc), using a ML-optimized base tree for likelihood calculations. ML phylogenies were reconstructed in PAUP* 4.0 beta 10 (Swofford, Citation2002). The rbcL ML analysis was performed using a tree-bisection–reconnection (TBR) heuristic search algorithm with 20 replicates of random taxon addition under the TIM1 + I + G model (Base frequencies: A = 0.3252 C = 0.1493 G = 0.1983 T = 0.3273; nst = 6; rate matrix: AC = 1.0000, AG = 4.1495, AT = 1.1925, CG = 1.1925, CT = 13.8085, GT = 1.0000; rates = gamma number of rate categories = 4, shape = 1.5790; and proportion of invariant sites = 0.5920). The cox2–3 ML analysis was performed using a TBR heuristic search algorithm with 20 replicates of random taxon addition under the TIM3 + I model (Base frequencies: A = 0.3351, C = 0.1454, G = 0.1467, T = 0.3729, nst = 6; rate matrix: AC = 4.8661, AG = 23.8553, AT = 1.0000, CG = 4.8661, CT = 39.7420, GT = 1.0000, rates = equal, and proportion of invariant sites = 0.6020). One hundred ML bootstrap (BS) replicates were conducted in Garli version 2.0 (Zwickl, Citation2006). For both data sets, the respective models were fixed and five replicates of random taxon addition were taken for every BS replicate. BI phylogenies were performed in MrBayes version 3.1.2 (Huelsenbeck & Ronquist, Citation2001; Ronquist & Huelsenbeck, Citation2003). In the rbcL data set, a GTR + I + G model was selected (lset nst = 6 rates = invgamma), the default priors were used, and MrBayes was run for 1 000 000 generations with a sample frequency of 1 tree for every 100 generations. The analysis was concluded when the average deviation of split frequencies was <0.01 (0.007423 exactly). One hundred and fifty trees were removed as burn-in, based on a plot of generations versus log probability. A majority-rule consensus tree containing posterior probabilities (PP) was constructed from the remaining trees. Similar methods were used to construct the cox2–3 BI phylogeny with the following exceptions: (1) a GTR + I model was used (lset nst = 6 rates = propinv), (2) the final average deviation of split frequencies was 0.003834, and (3) 50 trees were removed as burn-in, based on a plot of generations versus log probability.
ML and BI analyses were performed on a concatenated rbcL/cox2–3 dataset using Garli and MrBayes, respectively. ML and BI analyses were partitioned by locus. The ML model settings for rbcL partition were datatype = nucleotide, ratematrix = 6rate, statefrequencies = estimate, ratehetmodel = gamma, numratecats = 4, invariantsites = estimate. The ML model settings for cox2–3 partition were datatype = nucleotide, ratematrix = 6rate, statefrequencies = estimate, ratehetmodel = none, numratecats = 1, and invariantsites = estimate. The ML heuristic search included 20 replicates of random taxon addition. One hundred BS replicates were taken using the aforementioned parameters. Each BS replicate included 5 replicates of random taxon addition. The BI models for each partition were the same as the aforementioned single gene analyses. All parameters were unlinked and the rate multipliers for the two partitions were set to variable. MrBayes was run for 1 000 000 generations with a sample frequency of 1 tree for every 100 generations. The analysis was concluded when the average deviation of split frequencies was <0.01 (0.007525 exactly). One hundred and fifty trees were removed as the ‘burn-in’ based on a plot of generations versus log probability. The ML combined tree is shown as a phylogram with BS and PP labelled on their respective nodes (supplementary material). Uncorrected pairwise distances were calculated in PAUP* for both the rbcL and the cox2–3 datasets. All alignments and their resulting trees are available for download at http://www.treebase.org/treebase-web/search/study/summary.html?id=11800.
Voucher specimens for each sample were either fixed in preservative or pressed on herbarium paper. Vouchers for the new specimens from this study have been deposited at the Bartley Herbarium, Ohio University (BHO) and University of Michigan Herbarium (MICH) or at the National Herbarium of New South Wales (NSW) ().
Results
The rbcL alignment contained a total of 1282 characters, with no indels, and comprised 934 invariant characters, 275 variable parsimony-informative characters, and 73 variable characters that were parsimony-uninformative. The rbcL phylogeny was rooted with the following outgroup taxa: Batrachospermum gelatinosum (Linnaeus) DeCandolle (GenBank accession numbers AF029141, EF375888), Lemanea borealis G.F. Atkinson (AF029149), L. fluviatilis (Linnaeus) C. Agardh (AF029150), L. fucina var. parva M.L. Vis & Sheath (AF029151), Paralemanea annulata (Kützing) M.L. Vis & Sheath (AF029153, DQ449029, GQ285124), P. catenata (Kützing) M.L. Vis & Sheath (AF029154), and Tuomeya americana (Kützing) Papenfuss (AF029159).
ML and BI phylogenies were constructed based on rbcL gene sequence data and exhibited the same tree topology; the ML phylogram (–ln likelihood = 5160.48287) is shown with support values (PP/BS) marked on the nodes (). Outgroup taxa were pruned from . The genus Sirodotia comprised two major clades. The first clade included the taxa S. delicatula, S. huillensis and S. aff. huillensis and was strongly supported (PP 1.0/BS 98). Sirodotia delicatula was inferred as the monophyletic (PP 1.0/BS 96) sister group to a clade of S. huillensis specimens (PP 1.0/BS 100). The specimen of S. aff. huillensis, collected in South Africa, was inferred as sister to the aforementioned clades. Uncorrected pairwise distances for the specimens in the S. aff. huillensis–S. delicatula–S. huillensis clade ranged from 0 to 0.0282. The second major clade within the genus Sirodotia comprised specimens of S. suecica, S. tenuissima and S. goebelii. This clade was weakly supported (PP 0.83/BS<50). Uncorrected pairwise distances for taxa in this clade ranged from 0 to 0.0182 and the branch lengths within this clade were relatively short.
Fig. 1. Maximum likelihood (ML) tree (–ln likelihood = 5163.68597) based on rbcL DNA sequences, showing the relationships among specimens of Sirodotia species. The outgroup taxa have been pruned for presentation. Support values at nodes are ML bootstrap/Bayesian posterior probabilities: values <70% or 0.70 are not shown. Specimen information in .
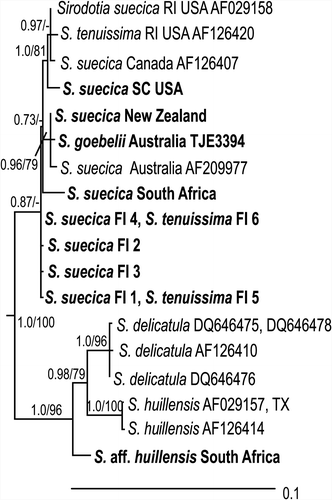
Within the second major clade, none of the taxa, S. suecica, S. tenuissima and S. goebelii, were reciprocally monophyletic (). The species S. tenuissima was also inferred as non-monophyletic as the three specimens were variously related to S. suecica specimens. The specimen of S. goebelii from the presumed type locality was in a strongly supported monophyletic group with two S. suecica specimens from Australia and New Zealand (PP 0.96/BS 82).
The phylogeographical distribution of specimens within the S. suecica–tenuissima–goebelii clade was investigated with the more variable cox2–3 spacer marker. The cox2–3 alignment contained 378 total characters and gaps were placed in the outgroup sequences. This alignment contained 298 invariant characters, 64 parsimony-informative characters, and 16 variable characters that were parsimony-uninformative and is available for download at http://www.treebase.org/treebase-web/search/study/summary.html?id=11800.
The cox2–3 ML and BI phylogenies exhibited similar tree topologies. The ML analysis resulted in three trees, each with a likelihood score of −ln 985.86775. These trees can be downloaded at http://www.treebase.org/treebase-web/search/study/summary.html?id=11800.
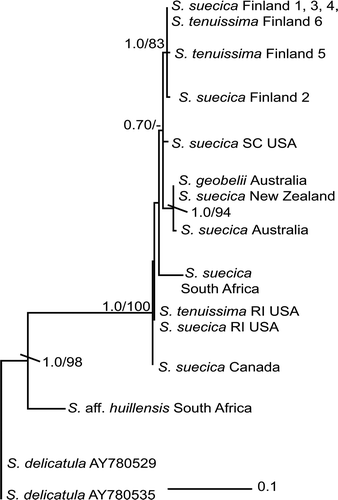
One of the three ML trees is represented as a phylogram with support values (PP/BS) in . As in the rbcL analyses, the S. suecica–S. tenuissima–S. goebelii clade was inferred as monophyletic, but with strong nodal support (PP 1.0/BS 100) in the cox2–3 phylogram. Uncorrected pairwise distances for taxa in this clade ranged from 0 to 0.0476 with very few base-pair substitutions among specimens. The Finland specimens (1–6) formed a clade (PP 0.86/BS 80). As in the rbcL analysis, S. goebelii from Australia formed a robustly supported clade with S. suecica species collected from Australia and New Zealand (PP 0.99/BS 98). Specimens from Canada, USA and South Africa formed an unsupported grade ().
Fig. 2. Maximum likelihood (ML) tree (–ln likelihood = 985.86775) based on cox2–3 spacer DNA sequences, showing the relationships among specimens of Sirodotia suecica. The specimens of S. delicatula, S. huillensis and S. aff. huillensis comprise the outgroup. Support values at nodes are ML bootstrap/Bayesian posterior probabilities, with values <70% or 0.70 not shown. Specimen information in .
We also concatenated the rbcL and cox2–3 sequences and the combined phylogeny is shown in the SupplementaryFigure. The results of the combined analysis were mostly similar to the single-gene analyses. The genus Sirodotia comprised two major clades. The first major clade included the taxa S. delicatula, S. huillensis and S. aff. huillensis. This clade was strongly supported (PP 1.0/BS 96). Sirodotia delicatula was inferred as the monophyletic (PP 1.0/BS 95) sister group to the specimen S. aff. huillensis collected in South Africa (PP < 0.70/BS < 70). Four specimens of S. huillensis (Arizona USA, Texas USA, GenBank AF029175, and GenBank AF126414) formed a well-supported monophyletic group (PP 1.0/BS 100) that was sister to the S. aff. huillensis–S. delicatula clade. The second major clade within the genus Sirodotia comprised S. suecica, S. tenuissima and S. goebelii. This clade was weakly supported (PP < 0.70/BS < 70). Within this clade, S. goebelii Australia, S. suecica New Zealand, and S. suecica GenBank AF209977 from Australia formed a strongly supported monophyletic group (PP 0.99/BS 88).
The specimen referred to here as S. aff. huillensis, could not be identified conclusively based on morphology, due to the limited diagnostic features present (). As in most Sirodotia species, there were obconic whorls on the main axis () and more barrel-shaped whorls of the smaller branches (). The specimen was monoecious with spermatangia at the fascicle tips (). The carpogonia had trichogynes with various morphologies () and no trichogynes with attached spermatia were observed. In some carpogonia, a protrusion of the base was not evident, but this may be due to their orientation (). No carposporophytes or carposporangia were seen.
Figs 3–8. Morphological features of Sirodotia aff. huillensis. 3. Main axis with obconic whorls (arrows). 4. Branches with appressed, barrel-shaped whorls (arrows). 5. Spermatangia (arrows) at fascicle tips. 6. Carpogonium with offset trichogyne (arrow). 7. Carpogonium with wavy trichogyne (arrow) and a symmetric base (double arrow). 8. Carpogonium with curved trichogyne (arrow). Scale bars = 150 µm (), 110 µm (), 10 µm () and 5 µm ().
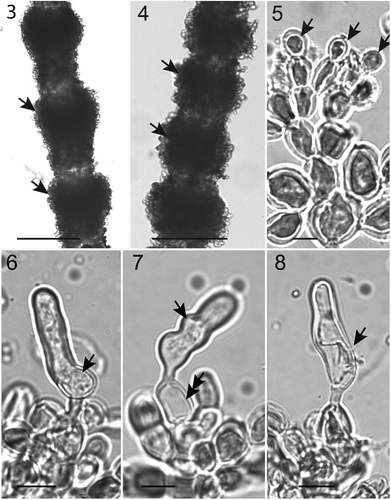
Table 2. Morphological characteristics of Sirodotia species recognized from molecular data. Some data taken from Kumano (Citation2002) and Necchi et al. (Citation2007) and references therein.
Discussion
The taxonomic status and classification of Sirodotia goebelii has been problematic because it was described from a rehydrated specimen from a single collection. Sirodotia goebelii was separated from S. suecica because it had generally pedicellate and deflexed trichogynes and long, profusely branched, erect gonimoblast filaments (Entwisle & Foard, Citation1999). In the material of S. goebelii examined at the time, the carpogonia had a broader base and the carposporangia were generally more elongate than in S. suecica (Entwisle & Foard, Citation1999). However, the molecular phylogenies (Figs , and the SupplementaryFigure) presented in this paper, using material from the presumed type locality in Victoria, Australia (see Appendix 1, Supplementaryfile), showed this taxon to differ very little from specimens of S. suecica, particularly those from Australia and New Zealand. While the newly collected material of S. goebelii does exhibit the key diagnostic characters from the protologue (Entwisle & Foard, Citation1999), it also extends the morphological range of most characters so that they now overlap further with those of S. suecica. Based on the strong molecular similarities between the two taxa, we propose that S. goebelii be considered a synonym of S. suecica (the name with precedence).
The recognition of Sirodotia tenuissima as a distinct taxonomic entity has been debated in the literature. Vis & Sheath (Citation1999) analysed two specimens of S. suecica and one specimen of S. tenuissima from North America and found that S. suecica would be paraphyletic if S. tenuissima were recognized as a separate taxon. Other taxonomic studies have recognized S. tenuissima as a distinct species based on morphological distinctiveness: e.g. the presence of separated truncate-pyramidal whorls with long internodes and carpospores in patches (Kwandrans & Eloranta, Citation2002; Eloranta & Kwandrans, Citation2007, Eloranta et al. Citation2011). The three S. tenuissima specimens from the USA and Finland included in the present study were variously related to the ten specimens of S. suecica in the rbcL, cox2–3 and combined phylogenies, and showed few base-pair differences. Thus, although these two taxa can be distinguished based on morphology, our results corroborate the previous study of Vis and Sheath (Citation1999) that S. tenuissima is synonymous with S. suecica.
In order to examine phylogeographical distribution patterns within Sirodotia suecica (including S. tenuissima, and S. goebelii) we used the cox2–3 spacer, which has been employed successfully in previous studies (Zuccarello & West, Citation2003; Rueness, Citation2005; Andreakis et al., Citation2007). Although the specimens analysed were collected from four different continents, only those from Australia and New Zealand formed a monophyletic group, potentially suggesting limited dispersal between this area and other parts of the world. Collections from Finland, the eastern United States, South Africa and Canada were a non-monophyletic grade, in which no phylogeographical patterns were evident. The specimen of S. suecica collected from South Africa is on a relatively long branch and may represent potential endemism. However, further research is necessary to this support this hypothesis. It should be noted that for Sirodotia, like other freshwater red algae, little is known about how plants are transported from one river system to another. However, from the molecular data for S. suecica, it would appear that this taxon is quite capable of long-distance dispersal.
The cox2–3 spacer is a non-protein coding region in the mitochondrial genome and is potentially much more variable than the protein coding rbcL gene. Sirodotia suecica samples (including S. goebelii and S. tenuissima) were collected from four continents, but the pairwise distances (0–0.0476) show a relatively low amount of genetic diversity. In comparison, cox2–3 spacer distances of Batrachospermum helminthosum Bory collected from a single continent (North America) ranged up to 0.06 (Chiasson et al., Citation2003). Batrachospermum macrosporum Montagne collected from North and South America exhibited an even broader range, from 0.00 to 0.19 (Vis et al., Citation2008), as did B. arcuatum collected from Bulgaria, China, Hawaii, New Zealand, Taiwan and the USA (0.00–0.148: Vis et al., Citation2010). Therefore, in comparison to these Batrachospermum, S. suecica exhibits a low level of genetic variability across a wide geographical area. This result provides further evidence for the synonymy of S. tenuissima and S. goebelii with S. suecica. However, it is still unclear why this taxon should have a lower genetic variability throughout its distribution than other Batrachospermales, since it does not appear to be fundamentally different in its autecology or life history.
The present research added two new specimens of Sirodotia huillensis from Texas and Arizona to previous phylogenies (Necchi et al., Citation2007). The new Texas specimen had an identical rbcL sequence to the previously sequenced specimen from Texas (GenBank AF029175), potentially because both were from the Edwards aquifer. The Arizona specimen showed little genetic difference from the Texas and Mexico specimens. The four S. huillensis locations in North America are rivers in the desert chaparral, but it is unknown whether this species is restricted to this biome. Our results are similar to those of previous studies showing that S. huillensis and S. delicatula are genetically distinct and more closely related to each other than to S. suecica (Vis & Sheath, Citation1999; Necchi et al., Citation2007).
The specimen of ‘Sirodotia aff. huillensis’ collected from South Africa was inferred as sister to a clade composed of S. delicatula and S. huillensis in the rbcL phylogeny. This result suggests that S. aff. huillensis is an entity separate from S. suecica, S. delicatula and S. huillensis (from Mexico, Texas and Arizona). However, we have not designated this specimen as a new species for several reasons. The first is that the phylogenetic placement of S. aff. huillensis differs between the three inferred phylogenies (Figs and and SupplementaryFigure). Secondly, S. aff. huillensis was received as a desiccated gametophyte sample ready for DNA extraction without a morphological voucher and the rehydrated sample provided limited morphological characters. It had carpogonia 33.6–42.1 µm in length, which is within the size range of S. huillensis (37–53 µm) and not much larger than S. delicatula (25–30 µm), according to Kumano (Citation2002). Likewise, spermatangia were observed, but we could not determine if they were in dense clusters as is diagnostic for S. huillensis (Necchi et al., Citation2007). All other characters, like the gonimoblast filaments arising from the protuberant or non-protuberant side of the carpogonium, or the indeterminacy of gonimoblast filaments, could not be observed. Furthermore, this specimen lacked any morphological autapomorphies that could be used for species description (). The third reason is that the only currently available specimens and DNA data representing S. huillensis are from North America, but the type locality for this taxon is Angola in Africa (Kumano, Citation2002). Therefore, the specimen from South Africa could represent ‘true’ S. huillensis and the specimens from North America attributed to S. huillensis could be a new species. Lastly, the exact location within the Sabie Catchment from which the South African specimen was collected is unknown and cannot be further defined because the collector, Gerhard Strydom, has passed away. We could have chosen not to include this specimen in our publication, however we prefer to share what morphological and molecular data we have. We provide the DNA sequences (GenBank accession numbers JF344727 and JF344717) as well the herbarium press (MICH) made of the desiccated sample in the hope that future researchers will elucidate the taxonomy of this potentially new species.
Synonymy
Sirodotia suecica Kylin, Nova Acta R. Soc. Scient. Ups. Ser. 4, 3: 38, (1912).
HETEROTYPIC SYNONYM: Sirodotia goebelii Entwisle & Foard, Aust. Syst. Bot. 12: 610, , 4B.
tejp_a_645885_sup_23778618.zip
Download Zip (654.3 KB)Acknowledgements
The authors would like to acknowledge funding sources for field collection and laboratory research as follows: Ohio University Postdoctoral Fellowship Program, the National Science Foundation (USA) grant numbers DEB0235676 and DEB0936855, and grant No. N N304 285937 from the Polish Ministry of Science and Higher Education. The following people are thanked for specimen collection: Wayne Chiasson, Thomas Dempster, Graeme Ellis, Emily Hollingsworth, Emily Johnston and the late Gerhard Strydom. Wayne Chiasson and Eric Salomaki assisted in some of the laboratory work. We thank Vijayanand Nadella and the Ohio University Genomics Facility for their expedient sequencing of our DNA samples. Many thanks to the correspondents who helped TJE trace S. goebelii from Western Australia to Narbethong in Victoria, albeit leading to its demise as a taxonomic entity: Hannes Hertel (Botanische Staatssammlung, München), Roberta Cowan (Murdoch University, Perth), Alison Vaughan (Royal Botanic Gardens Melbourne) and Amy Robson (Foster, previously The Hermitage).
References
- Andreakis , N , Procaccini , G , Maggs , C and Kooistra , WHCF . 2007 . Phylogeography of the invasive seaweed Asparagopsis (Bonnemaisoniales, Rhodophyta) reveals cryptic diversity . Molecular Ecology , 16 : 2285 – 2299 .
- Balakrishnan , MS and Chaugule , BB . 1980 . “ Indian Batrachospermaceae ” . In Taxonomy of Algae , Edited by: Desikachary , T.V. and Rao , V.N. 223 – 245 . Chennai : University of Madras .
- Carmona , JJ , Montejano , GZ and Necchi , Jr, O. 2006 . The ecology and morphological characterization of gametophyte and ‘Chantransia’ stage of Sirodotia huillensis (Batrachospermales, Rhodophyta) from a stream in Central Mexico . Phycological Research , 54 : 108 – 115 .
- Chiasson , WB , Machesky , NJ and Vis , ML . 2003 . Phylogeography of a freshwater red alga, Batrachospermum helminthosum, in North America . Phycologia , 42 : 654 – 660 .
- Eloranta , P and Kwandrans , J . 2007 . Freshwater red algae (Rhodophyta). Identification guide to European taxa, particularly to those in Finland . Norrlinia , 15 : 1 – 103 .
- Eloranta , P , Kwandrans , J and Kusel-Fetzmann , E . 2011 . “ Rhodophyta and Phaeophyceae ” . In Süßwasserflora von Mitteleuropa , Edited by: Büdel , B. , Gärtner , G. , Krienitz , L. , Preisig , H.R. and Schagerl , M. Vol. 7 , Heidelberg and Berlin : Spektrum Akademischer Verlag .
- Entwisle , TJ and Foard , HJ . 1999 . Sirodotia (Batrachospermales, Rhodophyta) in Australia and New Zealand . Australian Systematic Botany , 12 : 605 – 613 .
- Flint , LH . 1948 . Studies of fresh-water red algae . Am. J. Bot. , 35 : 428 – 433 .
- uiry, M.D. & Guiry, G.M. (2011). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 15 February 2011
- Hu , H and Wei , Y . 2006 . The Freshwater Algae of China: Systematics, Taxonomy and Ecology , Beijing : Science Press .
- Israelson , G . 1942 . The freshwater Florideae of Sweden. Studies on their taxonomy, ecology, and distribution . Symbolae Botanicae Upsalienses , 6 : 1 – 134 .
- Kumano , S . 1982 . Development of carpogonium and taxonomy of six species of the genus Sirodotia, Rhodophyta, from Japan and West Malaysia . Botanical Magazine,Tokyo , 95 : 125 – 137 .
- Kumano , S . 2002 . Freshwater Red Algae of the World , Bristol : Biopress Ltd .
- Kwandrans , J and Eloranta , P . 2002 . Variation of the genus Sirodotia (Batrachospermales, Rhodophyta) in some Nordic waters . Verhandlungen des Internationalen Verein Limnologie , 28 : 1370 – 1372 .
- Kylin , H . 1912 . Studien über die schwedischen Arten der Gattungen Batrachospermum Roth und Sirodotia nov. gen . Nova Acta Regiae Societatis Scientiarum Upsaliensis , 3 : 1 – 40 . ser. 4
- Huelsenbeck , JP and Ronquist , F . 2001 . MRBAYES: Bayesian inference of phylogeny . Bioinformatics , 17 : 754 – 755 .
- Necchi , Jr, O. and Vis , ML . 2005 . Reproductive ecology of the freshwater red alga Batrachospermum delicatulum (Batrachospermales, Rhodophyta) in three tropical streams . Phycological Research , 53 : 194 – 200 .
- Necchi , Jr, O. , Sheath , RG and Cole , KM . 1993 . Distribution and systematics of the freshwater genus Sirodotia (Batrachospermales, Rhodophyta) in North America . Journal of Phycology , 29 : 236 – 243 .
- Necchi , Jr, O. , Branco , CCZ and Branco , LHZ . 1999 . Distribution of Rhodophyta in streams from São Paulo state, southeastern Brazil . Archiv für Hydrobiologie , 147 : 73 – 89 .
- Necchi , Jr, O. , Vis , ML and Oliveira , MC . 2007 . Phylogenetic relationship of Sirodotia species (Batrachospermales, Rhodophyta) in North and South America . Cryptogamie, Algologie , 28 : 117 – 127 .
- Posada , D . 2008 . jModelTest: Phylogenetic Model Averaging . Molecular Biology and Evolution , 25 : 1253 – 1256 .
- Ronquist , F and Huelsenbeck , JP . 2003 . MRBAYES 3: Bayesian Phylogenetic Inference under mixed models . Bioinformatics , 19 : 1572 – 1574 .
- Rueness , J . 2005 . Life history and molecular sequences of Gracilaria vermiculophylla (Gracilariales, Rhodophyta), a new introduction to European waters . Phycologia , 44 : 120 – 128 .
- Sheath , RG and Sherwood , AR . 2002 . “ Phylum Rhodophyta (Red Algae) ” . In The Freshwater Algal Flora of the British Isles , Edited by: John , D.M. , Whitton , B.A. and Brook , A.J. 123 – 143 . Cambridge : Cambridge University Press .
- Sheath , RG , Müller , KM , Colbo , MH and Cole , KM . 1996 . Incorporation of freshwater Rhodophyta into the cases of chironomid larvae (Chironomidae, Diptera) from North America . Journal of Phycology , 32 : 949 – 952 .
- Skuja , H . 1938 . Die Süsswasserrhodophyceen der Deutschen Limnologischen Sunda-Expedition . Archiv für Hydrobiologie , 15 : 603 – 637 .
- Starmach, K. (1977). Phaeophyta – Brunatnice, Rhodophyta – Krasnorosty. Flora Słodkowodna Polski, vol. 14. PWN, Warszawa, Kraków
- Swofford, D.L. (2002). PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0b10. Sinauer Associates, Sunderland, MA
- Umezaki , I . 1960 . On Sirodotia delicatula Skuja from Japan . Acta Phytotaxonomica Geobotanica , 18 : 208 – 214 .
- Vis , ML and Entwisle , TJ . 2000 . Insights into the phylogeny of the Batrachospermales (Rhodophyta) from rbcL sequence data of Australian taxa . Journal of Phycology , 36 : 1175 – 1182 .
- Vis , ML and Sheath , RG . 1999 . A molecular investigation of the systematic relationships of Sirodotia species (Batrachospermales, Rhodophyta) in North America . Phycologia , 38 : 261 – 266 .
- Vis , ML , Saunders , GW , Sheath , RG , Dunse , K and Entwisle , TJ . 1998 . Phylogeny of the Batrachospermales (Rhodophyta) inferred from rbcL and 18S ribosomal DNA gene sequences . Journal of Phycology , 34 : 341 – 350 .
- Vis , ML , Hodge , JC and Necchi , Jr, O. 2008 . Phylogeography of Batrachospermum macrosporum (Batrachospermales, Rhodophyta) from North and South America . Journal of Phycology , 44 : 882 – 888 .
- Vis , ML , Feng , J , Chiasson , WB , Xie , S-L , Stancheva , R , Entwisle , TJ , Chou , J-Y and Wang , W-L . 2010 . Investigation of the molecular and morphological variability in Batrachospermum arcuatum (Batrachospermales, Rhodophyta) from geographically distant locations . Phycologia , 49 : 545 – 553 .
- West , W and West , GS . 1897 . Welwitsch's African freshwater algae . Journal of Botany , 35 : 1 – 7 . 33–42, 77–89, 113–122, 172–183, 235–243, 264–272, 287–304
- Zuccarello , GC and West , JA . 2003 . Multiple cryptic species: molecular diversity and reproductive isolation in the Bostrychia radicans/B. moritziana complex (Rhodomelaceae, Rhodophyta) with focus on North American isolates . Journal of Phycology , 39 : 948 – 959 .
- Zuccarello , GC , Burger , G and West , JA . 1999 . A mitochondria marker for red algal intraspecific relationships . Molecular Ecology , 8 : 1443 – 1447 .
- Zwickl, D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin
Supplementary material
The following supplementary material is available for this article, accessible via the Supplementary Content tab on the article's online page at http://dx.doi.org/10.1080/09670262.2011.645885.
TEJP 645885 supplementary material.docx