Abstract
Roystonea regia is a popular landscape palm famous for its smoothly sculpted white trunk, but sometimes it is spoiled by epiphytic green algae inhabiting the bark. We identified a trebouxiophycean green alga as the single dominant species growing on the trunk of a royal palm in Hainan, China. Light and transmission electron microscopy showed adult cells to be spherical with a diameter of 6.4–13.7 μm, and to contain a single cup-shaped parietal chloroplast. Each chloroplast contained a pyrenoid of 1.4–2.1 μm in diameter. The pyrenoid was penetrated by radially arranged tubular invaginations surrounded by pyrenoglobuli, which is similar to Heveochlorella hainangensis. Phylogenetic analyses inferred from nuclear-encoded 18S rRNA sequences placed strain ITBB A3-8 in the Watanabea clade (Trebouxiophyceae, Chlorophyta), closely related to H. hainangensis. But strain ITBB A3-8 differs sufficiently from H. hainangensis in significant aspects of ultrastructure, physiology, and nuclear and plastid ribosomal RNA and ITS sequences to be treated as a novel species, H. roystonensis Shuai Ma, V. Huss, Xuepiao Sun & Jiaming Zhang, sp. nov. The growth of the epiphytic H. roystonensis seems to be promoted by air pollution in the city, while the endophytic H. hainangensis requires an organic carbon source for optimal growth. Thus in the genus Heveochlorella both an epiphytic and an endophytic lifestyle of tree-dwelling green algae can be observed.
Introduction
Algae inhabit a wide variety of ecosystems including marine, freshwater, terrestrial and even extreme environments (Huss et al., Citation2002; La Rocca et al., Citation2009). Some microalgae have established symbiotic associations with other organisms, such as Chlorella with green hydra and Paramecium bursaria (Kovačević et al., Citation2005, 2007; Kodama et al., Citation2007), in which the algae provide energy and even detoxification to the hosts (Pardy, Citation1976; Reisser, 1976; Kovačević et al., Citation2005). Green algae inhabiting trees have been described in increasing numbers in recent years, either in lichen consortia, or free-living on the surface of bamboo or tree trunks (Hanagata et al., Citation1997; Rindi et al., Citation2006; Eliáš et al., 2010; Neustupa et al., 2011). Some Chlorella-like algae have even been found to be endophytic in plants, as shown for ginkgo and rubber trees (Trémouillaux-Guiller et al., Citation2002; Zhang et al., Citation2008). In Ginkgo biloba, for example, a eukaryotic alga resides inside the cells of this ‘fossil’ plant as a ‘precursor’ form, and is distributed worldwide by vertical inheritance (Trémouillaux-Guiller & Huss, Citation2007).
The royal palm (Roystonea regia) is a popular ornamental in tropical countries and was introduced into China from India and Sri Lanka. This palm is famous for its massive, symmetrical, smoothly sculpted and missile-like white trunk, which has an almost artificial appearance and makes a memorable impression on millions of tourists in Hainan each year. However, in recent years, some of these palm trees have become densely colonized by green algae, possibly due to the rapid increase of air pollutants by cars, making the trunk green and rough and badly affecting the aesthetic value of the trees. We took several algal isolates into culture from trunks and sought to characterize and identify them using morphological and molecular methods. We made an especially close comparison between the royal palm epiphytes and Heveochlorella hainangensis, previously isolated from rubber trees (Zhang et al., Citation2008).
Materials and methods
Field sampling and isolation of the algae
Field samples were collected from the bark of a royal palm in Haikou (N 19° 58′57″, E 110° 19′ 37″), Hainan, China. The samples were suspended in sterilized distilled water, and allowed to stand for a few minutes. The supernatant was sequentially diluted and plated on Tris-Acetate-Phosphate (TAP) medium (Harris, Citation1989), and incubated at 25°C, under 12 h light per day with a light intensity of 100 µmol photons m−2 s−1 from cool-white fluorescent tubes. The resulting single cell clones were subcultured on TAP medium until the morphology of the clones was uniform and no contamination detectable. The algal strains have been deposited in the Microorganism Collection Center at the Institute of Tropical Bioscience and Biotechnology, Hainan, and cultures stored at −80°C in 15% glycerol. Liquid TAP medium was used for suspension cultures, which were agitated at 120 rpm on a ZHWY-2000 orbital shaker (Shanghai Zhicheng, China). Incubation conditions were the same as above. A clonal algal strain ITBB A3-8 was further studied in this paper.
Microscopy
A small piece of bark with green algae was removed from a royal palm tree and sliced into thin sections. The sections were put on glass slides and observed with a Nikon 80i light microscope (LM) (Nikon, Tokyo, Japan). Cultured cells from different culture conditions were mounted on glass slides, examined and photographed with a 100× oil immersion lens. Cell sizes were measured with a micrometre eyepiece. The observations were made either in full exponential phase, or in late stationary phase of growth.
For transmission electron microscopy (TEM), cultured cells were harvested by centrifugation at 2000 × g for 5 min, the pellet was covered by a little egg white, fixed with 2.5% glutaraldehyde dissolved in 0.2 M phosphate buffered saline (PBS buffer: 135 mM NaCl, 4.7 mM KCl, 10 mM Na2HPO4, 2mM NaH2PO4, pH 7.2) for 3 h at 4°C, rinsed three times in PBS buffer for 10 min each time, and post-fixed in 1% osmium tetroxide (aqueous solution) for 2 h at 4°C. The pellet was then rinsed again three times in PBS buffer and dehydrated by increasing concentrations of acetone (50%, 70%, 80%, 90% and 100%, 10 minutes each) at room temperature. The samples were then embedded in Spurr's resin (Spurr, Citation1969). Ultrathin sections were prepared with a A130373 ultramicrotome (Leica, Germany), mounted on carbon-coated formvar slotted grids, sequentially stained with uranyl acetate and lead citrate, and observed with a JEOL 1010 (Jeol, Tokyo, Japan).
DNA extraction, PCR amplification and sequencing
The cultured cells were collected by centrifugation at 2000 × g for 5 min. The pellets were homogenized with a mortar and pestle. DNA was extracted with a universal DNA isolation kit (TaKaRa, Dalian, China) following the manufacturer's instructions. Nuclear-encoded 18S small subunit (SSU) rDNA was amplified using the primers 5′-ACCTGGTTGATCCTGCCAGTAG-3′ and 5′-ACCTTGTTACGACTTCTCCTTCCTC-3′ (Zhang et al., Citation2008), which were designed according to the consensus sequence of the available 18S rRNA sequences of chlorophyte species. The amplification conditions were as follows: 5 min at 94°C, 30 cycles of 30 s at 94ºC, 30 s at 58ºC and 2 min at 72ºC, and a final 10 min extension step at 72ºC. The internal transcribed spacer region (ITS), including ITS1, 5.8S rDNA and ITS2, of H. roystonensis and H. hainangensis FGG01 was amplified under the same conditions, using the primer pair 5′-CTTTGTACACACCGCCCGTC-3′ and 5′-ATATGCTTAAGTTGAGCGGGT-3′, which binds to conserved parts of the 3′-region of the 18S rDNA and the 5′-region of the 28S rDNA, respectively. The chloroplast 16S SSU rRNA sequence was amplified using the primers 5′-AGAGTTYGATCCTGGCTCAGG-3′ and 5′-GTGATCCAYCCYCACCTTCCA G-3′ (Zhang et al., Citation2008). The amplification conditions were 5 min at 94°C, 30 cycles of 30 s at 94ºC, 30 s at 55ºC and 1.5 min at 72ºC, and a final 10 min extension step at 72ºC. The oligonucleotides were synthesized by TaKaRa, Dalian, China. PCR products were cleaned with a DNA cleaning kit (TaKaRa, Dalian, China) and sequenced by Shangon Biotechnologies (Shanghai, China). The sequences were analysed with MacVector® 7.1 (Accelrys, Pharmacopeia, USA) and deposited in the GenBank database under accession numbers JN003600, JN003601, JX290371 and JX290372.
Phylogenetic analyses
The nuclear SSU rRNA gene sequences were included in large alignments of several hundred trebouxiophycean gene sequences including the most similar sequences as determined by BLAST searches, and the chloroplast SSU rRNA sequences were aligned with most available chlorophyte chloroplast SSU gene sequences. The alignment was carried out manually on a MicroVAX computer using the sequence editor program distributed by G. Olsen (Olsen et al., Citation1992) and with reference to secondary structure, determined according to the model of Wuyts et al. (2002). Highly variable regions that could not be aligned unambiguously for a subset of 38 relevant taxa in the 18S rDNA alignment were excluded from the analyses together with PCR primer binding regions and introns, resulting in a total of 1672 positions. Phylogenetic trees were inferred by the neighbour joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) methods using PAUP* 4.0b 10 (Swofford, Citation2002) for NJ and MP, and RAxML for the ML analyses (Stamatakis et al., 2008). The tree topology in was calculated by ML via the Web Server http://phylobench.vital-it.ch/raxml-bb/ using the CAT model for rate heterogeneity and choosing ‘Maximum Likelihood Search’ for the best scoring tree. 1000 bootstrap resamplings were calculated for NJ under the TrN+I+G model selected by Modeltest (Posada & Crandall, Citation1998; alignment in Supplementary File 1) and for MP choosing ‘New state’ for the treatment of gaps and random addition of sequences with each 10 replicates. For ML, 500 bootstraps were calculated using the same Web Server as indicated above.
The ITS-sequences were aligned using ClustalOmega (http://www.ebi.ac.uk/Tools/msa/clustalo/) with subsequent manual refinement of ITS2 by considering the conserved secondary structure of this region (Schultz et al., Citation2005; ). The phylogenetic analyses for a concatenated dataset of 18S+ITS in (2353 positions) were essentially done as described for the SSU rDNA sequences alone, except that GTR+I+G was selected by Modeltest as the best-fit model for the NJ bootstrap analysis (alignment in Supplementary File 2).
Results and discussion
Isolation of algal strains from royal palms
Growth of green algae on the trunk of the royal palm is common in Hainan and seems to be more vigorous in streets with heavy traffic (), compared to the growth in botanical gardens (), suggesting that the algae may obtain most of their nutrients from the air. Microscopical observation of bark samples () revealed that most of the algae were unicellular and located on the surface of the bark as clones independent from fungi (), although some alga–fungus consortia were also observed. Twelve pure algal strains were isolated, all of which were morphologically identical. Their 18S rDNA was partially sequenced and no variation found; thus all were considered to belong to a single dominant species among the epiphytic algal species dwelling on the palm. One strain (ITBB A3-8) was studied in detail.
Figs 1. Colonization of the algae on the royal palm (Roystonea regia). 1. Palms with algae growing vigorously on the trunk in a busy street in Haikou, Hainan Province, China. 2. Palms with poor algal growth in the Xinglong Tropical Botanical Garden, CATAS, Wanning County, Hainan. 3. Close-up, showing algae on the bark. 4. Bark section showing algae on the surface.

Morphological observations
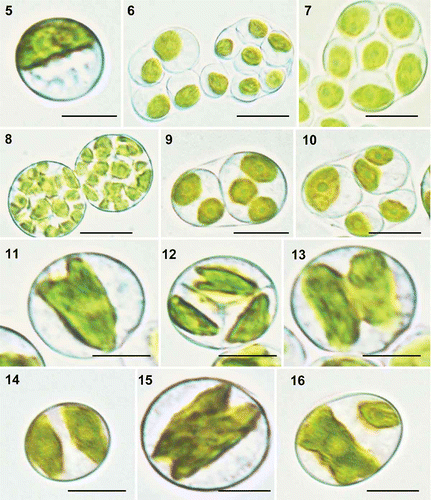
Strain ITBB A3-8 grew best at 28°C in TAP liquid medium at pH 7.0. Most of the adult cells were spherical, 6.4–13.7 μm in diameter, with a mean diameter of 9.8 μm, and contained a single parietal and cup-shaped chloroplast (). Autosporulation was the sole mode of reproduction. Most sporangia contained 4–16 autospores (, ) but a few contained 32 or more (). Sporangia with only two autospores were rarely observed, probably due to rapid division of the autospores; each of the two autospores usually contained two to three chloroplasts, ready for secondary division (). The autospores were ovoid, 6–9 μm in diameter, with a single ovoid chloroplast. The autospores usually divided equally, but in a few cases they differed in size due to unsynchronized development and division (, ), as deduced by the different numbers and sizes of chloroplasts in the autospores. The division pattern of the chloroplast was similar to that of Heveochlorella hainangensis FGG01, an alga isolated from rubber trees (Zhang et al., Citation2008), in that it was usually initiated from one end of the chloroplast and progressed longitudinally to the other end (). The two daughter chloroplasts then moved to opposite sides of the cell, becoming temporarily spindle-shaped (). Sometimes, the longitudinal division was initiated from both ends (), and in a few cases the chloroplast divided unequally ().
Figs 2. Heveochlorella roystonensis, strain ITBB A3-8, LM. 5. Mature vegetative cell with a parietal cup-shaped chloroplast. 6. Sporangia with four or eight autospores. 7. Sporangium with eight autospores. 8. Sporangia with 32 autospores. 9. Sporangium with two autospores, each containing two or three chloroplasts. 10. Sporangium with four autospores of different sizes. 11–13. Dividing chloroplasts. 14. Cell after completion of chloroplast division. 15. Chloroplast dividing from both ends. 16. Unequal division of a chloroplast. Scale bar = 5 μm.
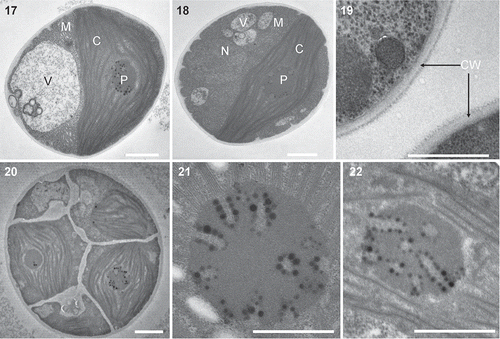
TEM micrographs revealed that the cell wall of strain ITBB A3-8 is smooth and double-layered, without any mucilage (). A cup-shaped parietal chloroplast accounted for approximately half of the volume of mature cells (). A pyrenoid, 1.4–2.1 μm in diameter, was located in the centre of the chloroplasts of both the mature cells (, ) and the autospores (). The stroma thylakoids were connected to the surface of the pyrenoid and radiated from it (, ). The pyrenoids were penetrated by radial, tubular invaginations 70–120 nm in diameter, with a central rod-like or fibrillar material and many surrounding pyrenoglobuli. This kind of pyrenoid ultrastructure is again similar to that of Heveochorella hainangensis (Zhang et al., Citation2008).
Figs 3. Heveochlorella roystonensis, strain ITBB A3-8, TEM. 17, 18. Mature vegetative cell showing the cup-shaped parietal chloroplast, pyrenoid, nucleus and mitochondria. 19. Two-layered cell wall of mature cell. 20. Sporangium containing multiple autospores. 21, 22. Ultrastructure of pyrenoids showing tube-like invaginations arranged radially. C, chloroplast; M, mitochondria; N, nucleus; V, vacuole; CW, cell wall. Scale bars = 1 μm.
Molecular characterization
In order to determine the phylogenetic position of strain ITBB A3-8, its nuclear-encoded 18S rDNA together with the adjacent ITS, as well as the chloroplast-encoded 16S rDNA were sequenced. The sequenced lengths of the 16S rDNA and the ITS were 1488 bp and 650 bp, respectively. The ITS of H. hainangensis, which was determined for comparison, had a length of 612 bp. The sequenced length of the 18S rDNA was 2500 bp including two introns. The two introns (353 bp and 382 bp) were located at positions 562 and 943 (E. coli numbering), which are well-known positions for group I introns (Cannone et al., Citation2002). The putative secondary structures of the two introns (not shown) were typical for group I introns of subgroup IC1 (Cannone et al., Citation2002), as in H. hainangensis, although their primary sequences were divergent.
Blast searches in the GenBank database using 605 nucleotides of the 18S rRNA sequence (positions 561–1165), in which most trebouxiophytes have no intron insertion site, returned highest similarities of 99% to ‘Chlorella’ luteoviridis strain MES A5-4 (AB006045), followed by 97% similarity to Heveochlorella hainangensis (EF595524), 96% to ‘Chlorella’ sp. MBIC10057 (AB058305), and 94% to several ‘Chlorella’ species including Heterochlorella luteoviridis (Neustupa et al., 2009). Excluding the intron regions is preferred over using the complete 18S rDNA sequence in blast searches, since group I introns are common among green algae and are fast evolving and mobile elements; however, they could not be excluded from reference strains during the blast search, thereby hiding a high similarity of the 18S exons. The chloroplast 16S rDNA was most similar to Heveochlorella hainangensis (95.3%, EF595524), but many fewer reference sequences were available in the database compared with 18S rDNA.
Phylogenetic position of ITBB A3-8
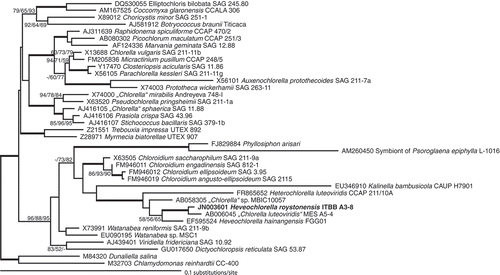
Phylogenetic trees were generated with 18S and 16S rRNA gene sequences and confirmed that strain ITBB A3-8 belongs to the Trebouxiophyceae. In the 18S rRNA tree, it was included in a clade also containing Heveochlorella hainangensis and ‘Chlorella’ luteoviridis strain MES A5-4 (). The Heterochlorella luteoviridis strains SAG 211-2a (Neustupa et al., 2009), CCAP 211/3, CCAP 211/4, CCAP 211/10A, CCAP 211/10E and another ‘Chlorella’ luteoviridis strain (MBIC 10057) formed a close sister clade (only one representative strain of H. luteoviridis, CCAP 211/10A, was included in the analysis, as all other strains had essentially the same sequence). The separation of Heveochlorella and Heterochlorella was not robustly supported in the phylogenetic analysis and the lack of a critical standard to combine or separate two genera (contrast the compensatory base changes in ITS2 used a criterion to judge conspecificity: see below) makes it difficult to decide whether these two genera should be kept apart or combined into a single genus Heveochlorella (since Heveochlorella was officially described earlier than Heterochlorella). More evidence and more strains from this lineage are needed to resolve this problem. However, the separation of the two genera is well supported by ultrastructural observations, since both H. hainangensis (Zhang et al., Citation2008) and strain ITBB A3-8 (, ) have tube-like thylakoids in the pyrenoid matrix and the pyrenoid is not surrounded by a starch sheath, whereas Heterochlorella luteoviridis strain SAG 211-2a has a starch sheath around the pyrenoid and the pyrenoid matrix is bisected by double-layer thylakoids (Neustupa et al., 2009). Unfortunately, the pyrenoid ultrastructure of ‘Chlorella’ luteoviridis MES A5-4, the closest relative of strain ITBB A3-8, could not be determined, as this strain was not available from any public alga collection, nor was any information about this strain known, including its habitat. The same applies to ‘Chlorella’ sp. MBIC 10057 which was also not available from public alga collections. The recently described genus and species Kalinella bambusicola (Neustupa et al., 2009) was the next most closely related to the Heveochlorella/Heterochlorella lineage in our phylogeny, although clearly divergent in its 18S rDNA. Its pyrenoid is surrounded by separate starch grains and bisected by many parallel thylakoids (Neustupa et al., 2009). A morphological comparison of the described taxa most closely related to ITBB A3-8, including pyrenoid ultrastructure as a useful taxonomic marker (Ikeda & Takeda, Citation1995), is given in .
Table 1. Morphological comparison between Heveochlorella roystonensis ITBB A3-8 and three closely related strains, Heveochlorella hainangensis FGG01, Kalinella bambusicola CAUP H7901 and Heterochlorella luteoviridis SAG 211-2a
Figs 4. Phylogenetic tree deduced from nuclear 18S rDNA sequences of Heveochlorella roystonensis and related trebouxiophycean green algae. The tree was rooted with two chlorophycean sequences as outgroups. Tree topology is based on a maximum likelihood (ML) analysis, which was mostly consistent with trees calculated by maximum parsimony (MP) and neighbour joining (NJ). The numbers at nodes indicate bootstrap support for NJ/MP/ML (see Methods). Statistical support of more than 90% for all three methods is indicated by a thick line. Bootstrap values lower than 50% are not shown. Branch lengths reflect the evolutionary distances indicated by the scale. GenBank accession numbers precede the taxon names.
Figs 5. Phylogenetic tree based on concatenated 18S+ITS sequence data of Heveochlorella and Heterochlorella with two more remotely related trebouxiophycean green algae as outgroups. A maximum likelihood analysis was used to infer the tree topology. The bootstrap support for NJ/MP/ML is indicated at the nodes (see Methods). Branch lengths reflect the evolutionary distances indicated by the scale. GenBank accession numbers precede the taxon names.

The chloroplast 16S rDNA sequences of strain ITBB A3-8 and Heveochlorella hainangensis clustered together with 100% bootstrap support, but as complete 16S sequences of Heterochlorella chloroplasts are not yet available, the 16S rDNA phylogeny did not contribute much to the resolution of the Heveochlorella–Heterochlorella lineage (tree not shown). Instead, we determined the ITS sequences of ITBB A3-8 and H. hainangensis and compared them with the published ITS sequence of Heterochlorella luteoviridis CCAP 211/10A (FR865652) to test a sister relationship of H. hainangensis and ITBB A3-8, and to provide assurance that they are distinct species. The phylogenetic tree in was based on a concatenated dataset of 18S+ITS sequences, with the trebouxiophytes Watanabea reniformis and Dictyochloropsis reticulata used as outgroups. Bootstrap support for a sister relationship of both Heveochlorella species was now excellent for NJ, MP and ML.
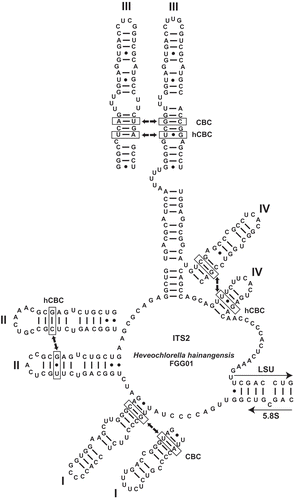
According to Mai & Coleman (Citation1997) and Coleman (Citation2007), two organisms should be considered as different species if the conserved parts of their ITS2 sequences differ by at least one compensatory base change (CBC). The ITS2 secondary structure model in for both Heveochlorella species shows that besides a considerable sequence variation (67.6% overall similarity for ITS2) there is one CBC at the conserved base of helix I, one hemi compensatory base change (hCBC) in helix II, one CBC and one hCBC in the conserved part of helix III, and at least one hCBC in the conserved part of helix IV. From this and all the other evidence given above, we conclude that ITBB A3-8 is a novel species of Heveochlorella, which we name H. roystonensis.
Figs 6. Secondary structure model of the internal transcribed spacer 2 (ITS2) of Heveochlorella hainangensis FGG01. Helices I, II, IV and the conserved part of helix III are shown for H. roystonensis ITBB A3-8 for comparison. Hemi-compensatory base changes (hCBCs) and full compensatory base changes (CBCs) are indicated for each helix.
Heveochlorella roystonensis Shuai Ma, V. Huss, Xuepiao Sun & Jiaming Zhang, sp. nov.
Figs 5–22
Descriptio: Algae microscopicae virides solitariae terricolae. Cellulae sphaericae, 6.4–13.7 μm diametro. Paries cellulae laevis bistratus. Chromatophora in cellulis iuvenalibus fusiformia, in cellulis adultis parietalia patelliformiaque, unoquoque pyrenoide unica 1.4–2.1 μm diametro praedito; pyrenoides a invaginationibus peripheralibus tubiformibus plusminusve radialibus penetrato; pyrenoglobuli invaginationes cingentes vel per matricem pyrenoidis dispersi. Propagatio per 4–16 autosporas globosas vel ellipsoideas.
Description: Green solitary microalgae, terrestrial. Cells spherical, 6.4–13.7 μm in diameter. Cell walls smooth and double-layered. Chloroplasts spindle shaped in young cells, parietal and cup-shaped in adult cells, with a single pyrenoid of 1.4–2.1 μm in diameter, which is penetrated by more or less radial, tubelike, peripheral invaginations; pyrenoglobuli surrounding the invaginations or scattered in the pyrenoid matrix. Reproduction by 4–16 spherical to ellipsoidal autospores.
Holotype: Deposited as ITBB A3-8 in the Microorganism Collection Center of the Institute of Tropical Bioscience and Biotechnology (ITBB), CATAS, Hainan, China and cryopreserved there in a metabolically inert state at −80°C.
Etymology: Derived from Roystonea regia, the royal palm.
Type locality: Hainan Province, China.
Distribution: Royal palms at the type locality, not known from elsewhere.
General discussion
The genus Chlorella was originally described as coccoid green algae surrounded by a smooth cell wall and reproducing exclusively by asexual autosporulation (Shihira & Krauss, Citation1965; Fott & Nováková, Citation1969). About 50 species were once assigned to this genus with traditional morphological methods (Fott & Nováková, Citation1969), including Chlorella luteoviridis. However, the lack of distinctive morphological features and the asexual reproduction of these algae caused considerable problems in taxonomic descriptions and classification (Kessler & Huss, Citation1992). In a biochemical and phylogenetic study, Chlorella species were found to be distributed in two classes of green algae, the Trebouxiophyceae and the Chlorophyceae (Huss et al., Citation1999). From the 50 species previously assigned to Chlorella, only three species now remained there: C. vulgaris, C. lobophora and C. sorokiniana (Krienitz et al., Citation2004). Other Chlorella-like species have been reclassified into distinctive genera, such as Auxenochlorella (Kalina & Punčochářová, Citation1987), Parachlorella (Krienitz et al., Citation2004), Chloroidium and Pseudochlorella (Darienko et al., Citation2010), Heveochlorella (Zhang et al., Citation2008) and Heterochlorella (Neustupa et al., 2009). However, there are still a number of Chlorella-like species not officially reclassified.
Heveochlorella roystonensis generally resembles H. hainangensis in morphology and ultrastructure, having a similar cell wall and chloroplast division pattern, and a pyrenoid with tube-like radial invaginations. But their differences are also significant: H. roystonensis grows well without sugar supplementation and has a single large pyrenoid, while H. hainangensis has several small pyrenoids and requires sugar supplementation for optimal growth (Zhang et al., Citation2008). In turn, the pyrenoid structure of both strains differs from that in their phylogenetically close relative Heterochlorella luteoviridis SAG 211-2a, whose pyrenoid is surrounded by a starch sheath and whose pyrenoid matrix is bisected by double-layer thylakoids (Neustupa et al., 2009), allowing their morphological distinction on the basis of pyrenoid ultrastructure. Therefore, judging by the molecular phylogeny, the ultrastructure of the pyrenoid is a more important feature for classification than the mere number and size of pyrenoids.
Both Heveochlorella roystonensis and H. hainangensis belong to the former Chlorella luteoviridis lineage within the Watanabea clade, together with Kalinella and Chloroidium. Examples of symbiotic associations of species belonging to the Watanabea clade are numerous: strain HHG of Chlorella saccharophila (= Chloroidium saccharophilum; Darienko et al., Citation2010) was isolated as an endosymbiont from the foraminiferan Heterostegina depressa (Lee et al., Citation1982; Huss et al., Citation1987), strain CAUP H7901 of Kalinella bambusicola is an epiphyte of the bamboo genus Gigantochloa (Neustupa et al., 2009), and Phyllosiphon arisari is a parasite of several species of Araceae (Aboal & Werner, Citation2011). Furthermore, Heveochlorella hainangensis was isolated as an endophyte of rubber trees (Zhang et al., Citation2008). Our strains of Heveochlorella roystonensis, however, were isolated as epiphytes from the surface of the bark of royal palms. Its growth seems to be promoted by air pollution in the city and the abundance of green algae on tree bark has been reported elsewhere to be positively correlated with airborne nitrogen pollution (Poikolainen et al., Citation1998), as has lichen growth (Gombert et al., Citation2003). One of the differences between the two Heveochlorella species is that the epiphytic H. roystonensis grows well autotrophically in media without carbon supplements, while the growth of the endophytic H. hainangensis is considerably enhanced by supplying a carbon source to the medium, which might be regarded as an adaptation to its supposed endophytic lifestyle (Zhang et al., Citation2008). In the genus Heveochlorella we may therefore witness the evolution from an epiphytic into an endophytic lifestyle of tree-dwelling green algae.
Supplementary material
Download Zip (27.3 KB)Acknowledgements
This research was supported by the Chinese International Science and Technology Cooperation Program (2010DFA62040), Natural Science Foundation of Hainan Province, China (No. 309051) and National Nonprofit Institute Research Grant of CATAS-ITBB (ITBBZX2008-4-3).
References
- Aboal , M. and Werner , O. 2011 . Morphology, fine structure, life cycle and phylogenetic analysis of Phyllosiphon arisari, a siphonous parasitic green alga . European Journal of Phycology , 46 : 181 – 192 .
- Cannone , J.J. , Subramanian , S. , Schnare , M.N. , Collett , J.R. , D'souza , L.M. , Du , Y. , Feng , B. , Lin , N. , Madabusi , L.V. , Muller , K.M. , Pande , N. , Shang , Z. , Yu , N. and Gutell , R.R. 2002 . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs . BMC Bioinformatics , 3 : 2
- Coleman , A.W. 2007 . Pan-eukaryote ITS2 homologies revealed by RNA secondary structure . Nucleic Acids Research , 35 : 3322 – 3329 .
- Darienko , T. , Gustavs , L. , Mudimu , O. , Menendez , C.R. , Schumann , R. , Karsten , U. , Friedl , T. and Pröschold , T. 2010 . Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta) . European Journal of Phycology , 45 : 79 – 95 .
- Eliáš , M. , Němcová , Y. , Škaloud , P. , Neustupa , J. , Kaufnerová , V. and Šejnohová , L. 2010 . Hylodesmus singaporensis gen. et sp. nov., a new autosporic subaerial green alga (Scenedesmaceae, Chlorophyta) from Singapore . International Journal of Systematic and Evolutionary Microbiology , 60 : 1224 – 1235 .
- Fott , B. and Nováková , M. 1969 . “ A monograph on the genus Chlorella. The fresh water species ” . In Studies in phycology , Edited by: Fott , B. 10 – 74 . Academia, Prague .
- Gombert , S. , Asta , J. and Seaward , M.R.D. 2003 . Correlation between the nitrogen concentration of two epiphytic lichens and the traffic density in an urban area . Environmental Pollution , 123 : 281 – 290 .
- Hanagata , N. , Karube , I. and Chihara , M. 1997 . Bark-inhabiting green algae in Japan (3) Chlorella trebouxioides and Ch. angusto ellipsoidea, sp. nov. (Chlorelloideae, Chlorellaceae, Chlorococcales) . Journal of Japanese Botany , 72 : 36 – 43 .
- Harris , E.H. 1989 . The Chlamydomonas sourcebook , San Diego : Academic Press .
- Huss , V.A.R. , Schwarzwalder , E. and Kessler , E. 1987 . Deoxyribonucleic acid reassociation in the taxonomy of the genus Chlorella. II. Chlorella saccharophila . Archives of Microbiology , 147 : 221 – 224 .
- Huss , V.A.R. , Frank , C. , Hartmann , E.C. , Hirmer , M. , Kloboucek , A. , Seidel , B.M. , Wenzeler , P. and Kessler , E. 1999 . Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta) . Journal of Phycology , 35 : 587 – 598 .
- Huss , V.A.R. , Ciniglia , C. , Cennamo , P. , Cozzolino , S. , Pinto , G. and Pollio , A. 2002 . Phylogenetic relationships and taxonomic position of Chlorella-like isolates from low pH environments (pH < 3.0) . BMC Evolutionary Biology , 2 : 13
- Ikeda , T. and Takeda , H. 1995 . Species-specific differences of pyrenoids in Chlorella (Chlorophyta) . Journal of Phycology , 31 : 813 – 818 .
- Kalina , T. and Punčochářová , M. 1987 . Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae) . Algological Studies , 45 : 473 – 521 .
- Kessler , E. and Huss , V.A.R. 1992 . Comparative physiology and biochemistry and taxonomic assignment of the Chlorella (Chlorophyceae) strains of the Culture Collection of the University of Texas at Austin . Journal of Phycology , 28 : 550 – 553 .
- Kodama , Y. , Nakahara , M. and Fujishima , M. 2007 . Symbiotic alga Chlorella vulgaris of the ciliate Paramecium bursaria shows temporary resistance to host lysosomal enzymes during the early infection process . Protoplasma , 230 : 61 – 67 .
- Kovačević , G. , Kalafatić , M. and Ljubešić , N. 2005 . Endosymbiotic alga from green hydra under the influence of cinoxacin . Folia Microbiologia Praha , 50 : 205 – 208 .
- Kovačević , G. , Kalafatić , M. and Ljubešić , N. 2007 . New observations on green hydra symbiosis . Folia Biologica Krakow , 55 : 77 – 79 .
- Krienitz , L. , Hegewald , E.H. , Hepperle , D. , Huss , V.A.R. , Rohr , T. and Wolf , M. 2004 . Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae) . Phycologia , 43 : 529 – 542 .
- La Rocca , N. , Andreoli , C. , Giacometti , G. , Rascio , N. and Moro , I. 2009 . Responses of the Antarctic microalga Koliella antarctica (Trebouxiophyceae, Chlorophyta) to cadmium contamination . Photosynthetica , 47 : 471 – 479 .
- Lee , J.J. , Reidy , J. and Kessler , E. 1982 . Symbiotic Chlorella species from larger Foraminifera . Botanica Marina , 25 : 171 – 176 .
- Mai , J.C. and Coleman , A.W. 1997 . The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants . Journal of Molecular Evolution , 44 : 258 – 271 .
- Neustupa , J. , Němcová , Y. , Eliáš , M. and Škaloud , P. 2009 . Kalinella bambusicola gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel coccoid Chlorella-like subaerial alga from Southeast Asia . Phycological Research , 57 : 159 – 169 .
- Neustupa , J , Eliáš , M. , Škaloud , P. , Němcová , Y. and Šejnohová , L. 2011 . Xylochloris irregularis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel subaerial coccoid green alga . Phycologia , 50 : 57 – 66 .
- Olsen , G.J. , Overbeek , R. , Larsen , N. , Marsh , T.L. , Mccaughey , M.J. , Maciukenas , M.A. , Kuan , W.-M. , Macke , T.J. , Xing , Y. and Woese , C.R. 1992 . The ribosomal database project . Nucleic Acids Research , 20 : 2199 – 2200 .
- Pardy , R.L. 1976 . The morphology of green hydra endosymbionts as influenced by host strain and host environment . Journal of Cell Science , 20 : 655 – 669 .
- Poikolainen , J. , Lippo , H. , Hongisto , M. , Kubin , E. , Mikkola , K. and Lindgren , M. 1998 . On the abundance of epiphytic green algae in relation to the nitrogen concentrations of biomonitors and nitrogen deposition in Finland . Environmental Pollution , 102 : 85 – 92 .
- Posada , D. and Crandall , K.A. 1998 . Modeltest: testing the model of DNA substitution . Bioinformatics , 14 : 817 – 818 .
- Reisser , W . 1976 . Die stoffwechselphysiologischen Beziehungen zwischen Paramecium bursaria Ehrbg. und Chlorella spec . in der Paramecium bursaria-Symbiose II. Symbiose-spezifische Merkmale der Stoffwechselphysiologie und der Cytologie des Symbioseverbandes und ihre Regulation. Archives of Microbiology , 111 : 161 – 170 .
- Rindi , F. , Lopez-Bautista , J.M. , Sherwood , A.R. and Guiry , M.D. 2006 . Morphology and phylogenetic position of Spongiochrysis hawaiiensis gen. et sp. nov., the first known terrestrial member of the order Cladophorales (Ulvophyceae, Chlorophyta) . International Journal of Systematic and Evolutionary Microbiology , 56 : 913 – 922 .
- Schultz , J. , Maisel , S. , Gerlach , D. , Müller , T. and Wolf , M. 2005 . A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota . RNA , 11 : 361 – 364 .
- Shihira , I. and Krauss , R.W. 1965 . Chlorella: physiology and taxonomy of forty-one isolates , Maryland : University of Maryland (College Park) .
- Spurr , A.R. 1969 . A low-viscosity epoxy resin embedding medium for electron microscopy . Journal of Ultrastructure Research , 26 : 31 – 43 .
- Stamatakis , A. , Hoover , P. and Rougemont , J. 2008 . A rapid bootstrap algorithm for the RAxML Web Servers . Systematic Biology , 57 : 758 – 771 .
- Swofford , D.L. 2002 . PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b 10. Sinauer Associates Sunderland, MA
- Trémouillaux-Guiller , J. and Huss , V.A.R. 2007 . A cryptic intracellular green alga in Ginkgo biloba: ribosomal DNA markers reveal worldwide distribution . Planta , 226 : 553 – 557 .
- Trémouillaux-Guiller , J. , Rohr , T. , Rohr , R. and Huss , V.A.R. 2002 . Discovery of an endophytic alga in Ginkgo biloba . American Journal of Botany , 89 : 727 – 733 .
- Wuyts , J. and Van de Peer , Y. 2002 . The European database on small subunit ribosomal RNA . Nucleic Acids Research , 30 : 183 – 185 . & de Wachter, R. (
- Zhang , J. , Huss , V.A.R. , Sun , X. , Chang , K. and Pang , D. 2008 . Morphology and phylogenetic position of a trebouxiophycean green alga (Chlorophyta) growing on the rubber tree, Hevea brasiliensis, with the description of a new genus and species . European Journal of Phycology , 43 : 185 – 193 .
Supplementary information
The following supplementary material is available for this article, accessible via the Supplementary Content tab on the article's online page at http://dx.doi.org/10.1080/09670262.2013.790996.Supplementary Files 1 and 2. Nexus files of the 18S rDNA and 18S+ITS alignments that were used to calculate the phylogenetic trees in and , respectively. The results of Modeltest evaluations for the best-fit models of DNA substitution (TrN+I+G and GTR+I+G, respectively) are included at the end of the files.