ABSTRACT
We describe Symbiodinium necroappetens sp. nov. found predominantly in diseased or thermally damaged tissues in some reef corals of the Greater Caribbean. Small, albeit fixed, differences in the ribosomal DNA (ITS2 and LSU) and cytochrome b (cob) indicate that S. necroappetens is evolutionarily separate, but closely related to S. microadriaticum (members of Clade A). However, haplotype sequences of the non-coding region of the psbA minicircle are highly divergent, signifying that the degree of genetic divergence between these sibling lineages is far greater than indicated by changes in rDNA. Small morphological differences also support the delineation of this species. The Kofoidian plate formula for S. necroappetens (x-plate, EAV, 4′, 5a, 8′′, 9-11s, 21c, 6′′′, 2′′′′, PE) is generally the same as described for S. microadriaticum, except for the number of cingulum plates (21 vs 22–24), but plate shapes and configurations differ. Nuclear and mitochondrial volumes calculated from ultrastructural serial sections (published previously) also distinguish it from S. microadriaticum and S. pilosum. There are significant physiological differences in the response of S. necroappetens to high pCO2 and thermal stress when compared with S. microadriaticum, indicating that large functional differences exist even among closely related species. This species appears to be necrotrophic rather than mutualistic. Before it was recognized as a distinct entity, reports on its ecology contributed to the supposition that members of Clade A Symbiodinium were opportunistic. Available evidence indicates that S. necroappetens exists at low environmental background levels, but may ‘proliferate’ selectively in artificial growth media, or emerge opportunistically in bleached coral colonies during early recovery from severe stress, or in diseased necrotic tissues, especially in colonies of the Orbicella (formerly Monstastraea) annularis complex. However, S. necroappetens fails to persist at detectable levels as populations of the typical symbiont recover. The description of this species raises awareness of the broad functional and ecological diversity exhibited by members of this large dinoflagellate genus.
INTRODUCTION
Dinoflagellates in the genus Symbiodinium, as mutualistic endosymbionts, are distributed almost everywhere that their metazoan and protistan hosts occur. Geographically, this includes all tropical and subtropical near-shore habitats as well as many temperate high-latitude environments. The environmental heterogeneity accompanying this broad geographic distribution, combined with the influences of hundreds of different host species, presents numerous opportunities for niche diversification, leading to speciation. Indeed, genetic evidence and ecological patterns indicate the evolution of an enormous diversity of Symbiodinium species via selection imposed by biotic and abiotic factors (LaJeunesse et al., Citation2014; Thornhill et al., Citation2014).
Phylogenetic analyses of Symbiodinium reveal multiple divergent lineages, referred to as clades (designated A, B, C … etc.), which differ in their relative ecological dominance depending on regional and local environmental factors (Baker, Citation2003; LaJeunesse et al., Citation2010). Some clades contain numerous ecologically distinct lineages that are resolved by fixed, albeit small, nucleotide differences in nuclear ribosomal DNA (involving comparisons of the most common intragenomic variant in an organism’s ribosomal array; sensu Thornhill et al., Citation2007; Sampayo et al., Citation2009) and sequences of the chloroplast ribosomal large subunit (cp23S, Santos et al., 2002). The majority of these genetic ‘types’ also exhibit restricted geographic distributions that correspond to ocean basins and latitude (LaJeunesse et al., Citation2010). Differences in niche and biogeography indicate that most types are independently evolving species lineages (LaJeunesse et al., Citation2012, Citation2014; Thornhill et al., Citation2014).
The detection and genetic analysis of Symbiodinium from water samples, sediments and various other substrata reveals the existence of unusual genetic lineages not detected in host tissues and signifies the existence of a diverse group of ‘free-living’, opportunistic and possibly non-mutualistic species (LaJeunesse, Citation2002; Hirose et al., 2008; Porto et al., Citation2008; Pochon et al., Citation2010; Jeong et al., Citation2014). These and other findings have led to the growing realization that the ecological breadth of the genus Symbiodinium extends well beyond endosymbiotic mutualisms (LaJeunesse, Citation2001, Citation2002; Lesser et al., Citation2013; Jeong et al., Citation2014). Some have presumed that members of Clade A (sensu lato) are stress tolerant, fugitive opportunists and even parasites (Toller et al., Citation2001; Stat et al., Citation2008; Lesser et al., Citation2013). This perception began when Rowan et al. (Citation1997) found that ‘Clade A’ increased its proportional representation following a natural bleaching event, and Toller et al. (Citation2001) reported that ‘Clade A’ was common in Orbicella (formerly Montastraea) samples taken during bleaching experiments and from tissues with yellow-band disease (YBD, sometimes called yellow-blotch disease). Although Toller et al. (Citation2001) recognized the potential for cryptic diversity within clades (both Rowan & Powers (Citation1991) and Toller et al. (Citation2001) acknowledged that the small subunit rDNA did not resolve species), they presumed, in the absence of evidence to the contrary, that their ‘Clade A’ entity was the same as the species most often found on the tops of shallow Orbicella colonies, currently known as Symbiodinium A3 (Warner et al., Citation2006; Kemp et al., Citation2008).
As the ancestral lineage of the genus, Clade A Symbiodinium is composed of species with broad differences in ecology (LaJeunesse et al., Citation2009a). Currently, there are more than 18 distinctive taxa (i.e. ‘types’ or ‘species’) in Clade A (reviewed in LaJeunesse et al., Citation2009a). Many of these exist as dominant, functionally important, symbionts in various animals (LaJeunesse et al., Citation2009a), including reef-building corals (Baums et al., Citation2014), other reef Cnidaria (fire corals, zooanthids, jellyfish, etc.; LaJeunesse, Citation2002; Finney et al., Citation2010), as well as molluscs in the subfamily Tridacninae (giant clams; Lee et al., Citation2015). Several Clade A types are prevalent in the Greater Caribbean (northwestern tropical Atlantic). For example, type A3 occurs predictably in many reef coral species including Acropora sp. (S. ‘fitti’ nomen nudum), Stephanocoenia and Orbicella. Type A4 and A4a are commonly found in colonies of the genus Porites spp. and with large anemones (Condylactis and Stichodactyla; Finney et al., Citation2010). Symbiodinium microadriaticum (Lee et al., Citation2015), also called type A1, occurs in the upside-down mangrove jellyfish Cassiopea xamachana (LaJeunesse, Citation2002). In addition, there are two formally described Symbiodinium spp. in Clade A that appear to exist as non-symbiotic free-living taxa. Symbiodinium pilosum (synonymous with type A2; Trench & Blank, Citation1987), grown from culture, has yet to be detected in analyses of field-collected host samples (e.g. LaJeunesse, Citation2002) and does not successfully infect experimental aposymbiotic hosts (LaJeunesse, Citation2001; Xiang et al., Citation2013). These observations indicate that some Symbiodinium belong to non-mutualistic functional guilds (LaJeunesse, Citation2002; Hansen & Daugbjerg, Citation2009; Lesser et al., Citation2013).
Another Symbiodinium whose ecological niche appears to differ from mutualistic species is that of type A13. It was originally characterized in culture and distinguished from S. microadriaticum (A1) by a single base substitution in the ITS2 (formerly listed as type A1.1 by LaJeunesse (Citation2001); ). For years it was assumed that it represented one of several enigmatic entities obtained during the culturing process, but for which there was no evidence of their ecological distribution in the environment (see above; LaJeunesse, Citation2002). Yet, A13 was eventually found in samples from severely stressed and bleached colonies of the reef-building coral, Orbicella spp., and in Porites spp. collected during the mass coral bleaching and mortality event in Barbados, 2005 (LaJeunesse et al., Citation2009b). This provided the first clues as to the ecological distinctiveness of type A13 and the need for its own taxonomic binomial.
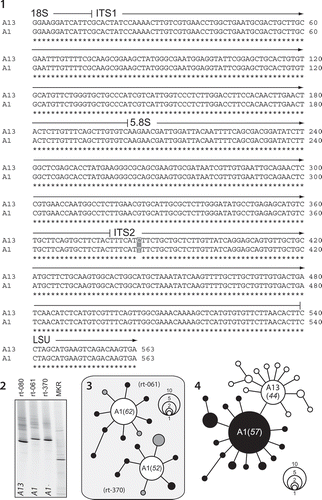
Figs. 1–4. The analysis of ITS rDNA from closely related Symbiodinium. Fig. 1. A single fixed nucleotide change across the entire ITS1-5.8S-ITS2 region differentiates S. microadriaticum (type A1 sensu LaJeunesse, Citation2001) from S. necroappetens (formerly, type A13, or A1.1). Fig. 2. The visual resolution of these entities using DGGE fingerprinting of ITS2 amplifications on cultured isolates. Fig. 3. The genomes of S. microadriaticum strains in culture share a dominant ITS2 sequence diagnostic of the species, but can differ in the content and identity of low abundance and derived sequence variants (viewed as spokes around the numerically dominant centre sequence in unrooted phylogenies). Grey shaded circles correspond to the rare sequence variants found in both CCMP 2464 (rt-061) and CCMP 2467 (rt-370). Fig. 4. The numerically dominant ITS2 sequence in the genome of S. necroappetens differs from S. microadriaticum by a single nucleotide transition. Rare intragenomic sequence variants (white circles) found in isolate CCMP 2469 (rt-080) are derivatives of the ‘A13’ sequence and do not overlap with sequence variants (black circles) found in the combined sequences recovered from CCMP 2464 and CCMP 2467. The number of times a particular sequence variant was cloned and sequenced is indicated by circle size. Additionally, the frequency (%) that the numerically dominant ITS2 variant was recovered is listed in parentheses.
Here, we describe Symbiodinium A13 as a new species that exhibits ecological opportunism in bleached or necrotic animal tissues, using genetic, morphological, physiological and ecological data. In addition to the analysis of several cultured isolates, this work includes the reanalysis of Orbicella samples with YBD and colonies recovering from induced low light (dark) stress originally examined by Toller et al. (Citation2001), as well as analyses of type A13 from an independent study examining induced thermal stress, bleaching and early recovery of Orbicella colonies from the Yucatan, Mexico (Grotolli et al., Citation2014). We also performed additional genetic analyses on collections made from bleached and recovering corals from Barbados during the 2005 Eastern Caribbean thermal heating event (LaJeunesse et al., Citation2009b). Finally, samples of YBD tissues obtained from the Florida Keys, Puerto Rico, and Barbados were analysed. Our defining of formal Symbiodinium species using independent lines of evidence serves a practical need to enhance research into coral-dinoflagellate symbioses by standardizing scientific communication and improving interpretations of ecological data (LaJeunesse, Citation2001, Citation2002; Sampayo et al., Citation2009; LaJeunesse et al., Citation2012, Citation2014; Stat et al., Citation2012).
MATERIALS AND METHODS
Field-collected samples
Specimens containing Symbiodinium type A13 that were collected, preserved and examined genetically came from several independent collections. Samples from Barbados were collected before (July and August), during (December) and approximately 6 months (May 2006) following the 2005 mass coral bleaching event (LaJeunesse et al., Citation2009b). Seven colonies of Orbicella with YBD were sampled from Turrumote Reef, La Parguera, Puerto Rico, in 2004 (17°56.097N, 67°01.130W) and included yellowed, transitional and healthy tissues. Healthy and yellowed tissues of six colonies of Orbicella from the Dry Tortugas (Sherwood Forest) were biopsied in August 2005. For colonies of Orbicella with YBD from Barbados (n = 10 samples, five colonies), the Florida Keys (n = 12 samples, six colonies), and Puerto Rico (n = 21 samples, seven colonies), one lesion was sampled per colony and a sample was taken from yellowed tissues and from a healthy zone approximately 10 cm away. For Puerto Rico, a third sample was taken from the bleached tissues in a zone between healthy and yellowed tissues. A subset of samples of YBD (n = 4) and experimentally bleached colonies (n = 6) of Orbicella from Panama originally analysed by Toller et al. (Citation2001) were also obtained and reanalysed to determine the Clade A identity with higher resolution genetic markers. Finally a select set of samples was obtained from a recent study investigating the response of Orbicella colonies exposed to repeated thermal stress and their symbiont composition monitored during recovery (see Grottoli et al., Citation2014 for details of experimental manipulation and sample collection).
Cultured isolates
Several cultured isolates identified as type A13 were independently obtained from different host origins. They included the giant Caribbean sea anemone, Condylactis gigantea (CCMP 2469, formerly rt-080; ‘rt’ is used to indicate original cultures from the Robert K. Trench collection) and the upside-down jellyfish, Cassiopea frondosa (culture rt-048), from Jamaica. Additional cultures of A13 were obtained from Mary Alice Coffroth (SUNY Buffalo) who isolated them from juveniles of the gorgonian sea fan, Plexaura kuna (cultures 225 and 708), collected from San Blas Island, Panama. Cultures of S. microadriaticum (= type A1; rt-061 and rt-370) were also used for comparative purposes in describing intragenomic variation of rDNA. All cultures were maintained in the artificial culture medium ASP-8A (Blank, Citation1987) at 26°C, illuminated by banks of full spectrum fluorescent bulbs delivering 80–120 µmol photons m−2 s−1 photosynthetically active radiation (PAR) on a 14:10 (light:dark) photoperiod. Cells were harvested by centrifugation at 800–1000 × g.
Genetic analyses
DNA extractions for most samples followed the protocol described by LaJeunesse et al. (Citation2003). All DNA samples from the investigations of Toller et al. (Citation2001) were extracted as per their protocols. All field samples were initially screened using denaturing gradient gel electrophoresis (DGGE) of ITS2 PCR amplifications (LaJeunesse & Trench, Citation2000). Bands diagnostic of the resulting ITS fingerprint were excised, eluted, re-amplified and directly sequenced as described by LaJeunesse (Citation2002). ITS2 amplifications from cultures rt-080, rt-061 and rt-370 were bacterially cloned using the pGEM-T cloning kit (Promega) following the manufacturer’s instructions and 13–16 cloned inserts sequenced to assess intragenomic variation of rDNA. The partial LSU (D1/D2 domains) chloroplast cp23S and mitochondrial cob genes were amplified and directly sequenced for all cultures and field collected samples according to Zardoya et al. (1995), Zhang et al. (Citation2000) and Zhang et al. (Citation2005), respectively. The primers psbAFor_1 (5′- GCA GCT CAT GGT TAT TTT GGT AGA C - 3′) and psbARev_1 (5′- AAT TCC CAT TCT CTA CCC ATC C - 3′), designed to have efficacy on most Symbiodinium (LaJeunesse and Thornhill, Citation2011), were used to amplify and sequence the non-coding region of the psbA minicircle (psbAncr). The PCR conditions for amplification of the psbAncr were: 94°C for 2 min; then 40 cycles of 94°C 10s, 55°C for 30 s and 72°C for 2 min; followed by a final extension at 72°C for 10 min. Direct Sanger sequencing on PCR amplified DNA was performed using Big Dye 3.1 reagents (Life Sciences) and the reaction products analysed on an Applied Biosystems 3730XL instrument.
Base calling on chromatograms were visually inspected for accuracy (Sequence Navigator) and the edited sequences aligned initially using the online application of ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Further adjustments to alignments were made upon visual inspection of the output file. Final edited sequences were deposited in GenBank. Phylogenetic analyses using Maximum Parsimony were conducted using the software PAUP (v. 4.0a136, Swofford, Citation2002) on aligned sequences. Bootstrap support of branching nodes was based on 1000 replicates. A phylogenetic reconstruction of the psbAncr was conducted using Bayesian Inference using MrBayes v3.2.1 (Huelsenbeck & Ronquist, Citation2001) implementing General Time Reversible (GTR)+I+ Γ (C3-group psbAncr data). Each MCMC analysis was run for 1.0×106 generations and sampled every 100 generations. The first 25% of trees were discarded as burn-in corresponding with the convergence of chains.
Scanning electron microscopy (SEM)
SEM was used to characterize cell size and the amphiesmal plate shape, number and arrangement for cells from culture CCMP2469 (rt-080). Cells from dense cultures were fixed for 10 min in osmium tetroxide at a final concentration of 0.3% (v/v) in seawater. Cell collection, dehydration, drying and mounting were performed as described in Kang et al. (Citation2010).
RESULTS
Species description
Symbiodinium necroappetens LaJeunesse, Lee, Knowlton & Jeong, sp. nov. (–)
DIAGNOSIS: Coccoid cell size ranges from 8–12 µm. The Kofoidian plate arrangement of motile cells consisted of x-plate, elongated amphiesmal vesicle (EAV), 4′, 5a, 8′′, 9-11s, 2 cingulum rows, 6′′′, and 2′′′′.
Nucleotide sequences of the large ribosomal subunit rDNA (GenBank accession no. KP114226), internal transcribed spacer rDNA (accession no. AF333504), partial chloroplast large sub-unit, cp23S (accession no. KP114225), mitochondrial cytochrome b, cob (accession no. KP114224), and partial sequences of the psbA and psbAncr (accession nos KP165611–KP165619) genetically define this species.
HOLOTYPE: The holotype of Symbiodinium necroappetens, culture CCMP2469 fixed with 0.3% (w/v) osmium tetroxide, was deposited in the US National Herbarium, National Museum of Natural History, Smithsonian Institution, Washington DC, USA, as US Algal Collection specimen # 222985.
TYPE LOCALITY: Discovery Bay, Jamaica (18°27′59′′N; 77°24′03′′W)
HABITAT: Probably present at low background concentrations in various shallow tropical cnidarians in the northwestern tropical Atlantic. It can occur in abundance with colonies of the Orbicella (formerly Montastraea) annularis complex recovering from severe stress or in the necrotic region of yellow-band disease.
ETYMOLOGY: From the Greek ‘necrosis’ and Latin ‘appetens’ meaning necrosis-desiring.
SYNONYMS: Symbiodinium microadriaticum subsp. ‘condylactis’ (Blank & Huss, Citation1989). Banaszak et al. (Citation1993) subsequently proposed that it was a separate species based on differences in iso-electric profiles of the peridinin-chlorophyll a-proteins and motility patterns, and proposed the informal designation S. ‘cariborum’ (nomen nudum). LaJeunesse (Citation2001) initially designated it as the genetic type A1.1 based on phylogenetic evidence, but subsequently changed it to type A13 (LaJeunesse et al., Citation2009a; see below).
Genetic identification of S. necroappetens from cultures and field samples
Isoclonal cultures of Symbiodinium necroappetens were genetically characterized using direct sequencing of the ITS1-5.8S-ITS2 nuclear rDNA, which found that a single nucleotide substitution in the ITS2 region distinguished it from S. microadriaticum (). LaJeunesse (Citation2001) named it A1.1 to distinguish it from type A1 (= S. microadriaticum). That alpha-numeric was later changed to type A13 because subsequent ecological evidence clearly differentiated it from S. microadriaticum (see below, LaJeunesse et al., Citation2009a). Analysis by DGGE of ITS2 amplifications shows that the genomes of S. necroappetens and S. microadriaticum are relatively homogenized and can be diagnosed by the sequence variant that is numerically dominant across the ribosomal array (). Analysis of intragenomic variation using bacterial-cloning/sequencing found that the individual genomes from each species contain some intragenomic variation with no apparent overlap, and that these variants are mutations of the sequence variant that is most abundant across the rDNA tandem array (, ).
Symbiodinium necroappetens was identified from samples and cultured isolates obtained from locations throughout the tropical northwest Atlantic (). It was detected at relatively high abundances in field-collected samples from coral tissues that were diseased () or severely bleached from natural conditions or experimental manipulations ().
Figs 5–7. The geographic and ecological distributions of S. necroappetens. Fig. 5. The known geographic distribution for Symbiodinium necroappetens sp. nov. Calendar years are indicated when samples or cultures of S. necroappetens were obtained from a particular location. Fig. 6. This species may occur at detectable densities in yellow-band (yellow-blotch) diseased tissues on colonies of the reef builder, Orbicella spp. (photo by Dustin Kemp). Fig. 7. The new species can also be found in colonies recovering from severe natural (e.g. Orbicella and Porites) and in colony fragments recovering from experimentally induced bleaching (photo by Hazel Oxenford).
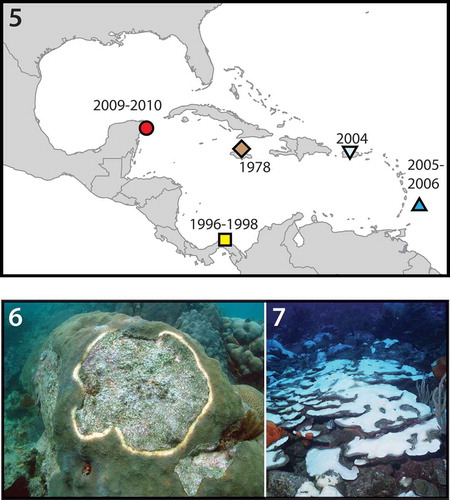
In December 2005, 6 weeks after cessation of prolonged thermal stress, when many corals were severely bleached, a total of 72 colonies comprising 10 scleractinian genera and the octocoral Erythropodium were sampled and the symbionts examined. From these samples, S. necroappetens was recovered in four out of 14 Orbicella spp. colonies (three sampled at 12–15 m and one at 2–4 m, all severely bleached), in one out of six colonies of Porites porites (specimen sampled at 2–4 m), and in one out of 14 colonies of Porites astreoides (specimen sampled at 12–15 m). These coral communities were sampled approximately 6 months later (May/April 2006); from the analysis of 34 samples comprising six scleractinian genera and the octocoral genus Erythropodium, S. necroappetens was identified in only one colony of Orbicella sp. (still pale and recovering) out of 21 Orbicella colonies sampled, suggesting that the ability to detect S. necroappetens at relatively high concentrations is ephemeral.
Symbiodinium necroappetens occurred in colonies of Orbicella sp. recovering from the controlled perturbation of normally functioning symbioses (experimental coral bleaching) conducted at two distant locations in Central America: Puerto Morelos, Mexico, and San Blas, Panama. Experiments conducted in 1997–1998 at Aguadargana reef, San Blas Archipelago, Republic of Panama, involved exposing colonies of Orbicella annularis and O. faveolata to severe low-light stress (Toller et al., Citation2001). Many samples from tissues recovering for 8–13 weeks from a 4–7 week ‘dark’ treatment, and explanted to a common coral nursery site at 9 m, contained S. necroappetens exclusively, or with other Symbiodinium spp. typically associated with Orbicella spp.
Symbiodinium necroappetens was found in the yellowed tissues adjacent to the exposed dead skeleton in YBD from Turrumote Reef, La Parguera, Puerto Rico (in six out of seven lesions originating from independent colonies) and Cayos Limones in the San Blas Archipelago, Panama (11 out of 12 colonies). However, S. necroappetens was not detected using PCR-DGGE in diseased tissues sampled from Barbados (July 2005) nor from the Florida Keys (August 2005).
Phylogenetic position of S. necroappetens within Clade A
Direct sequencing of ITS and LSU rDNA produced clean, unambiguous sequences. Sequences of the ITS2 and LSU each differed by a single base substitution from the ITS and LSU sequences diagnostic of S. microadriaticum (). Screening of ITS2 variation using DGGE showed a single prominent band in the gel () and indicated that, like S. microadriaticum, the ribosomal array of S. necroappetens possessed one numerically dominant variant (Arif et al., Citation2014). While S. necroappetens and S. microadriaticum are diagnosed by similar ITS rDNA sequences, bacterial cloning failed to discover the presence of the A13 sequence in the genomes of two strains of S. microadriaticum (rt-061, rt-370), nor the presence of the A1 sequence in a strain of S. necroappetens (rt-080, ). Instead, this method recovered a high proportion of cloned ITS2 sequences that matched the diagnostic sequence derived from direct sequencing and DGGE screening (–). Additionally, 50–60% of bacterially cloned sequences were variants that differed from the most commonly recovered sequence by one or two base substitutions (, ).
The cytochrome b (cob) sequences differed by five nucleotide substitutions (two synonymous and three non-synonymous). The cp23S chloroplast marker, used commonly to delimit different Symbiodinium, did not distinguish S. necroappetens from S. microadriaticum. Sequences of cp23S were identical from the cultures and samples of each species obtained from locations across the Caribbean. The concatenation and phylogenetic analysis of nuclear rDNA with mitochondrial and chloroplast gene sequences unambiguously differentiated these species ().
Figs 8–9. The phylogenetic analyses of nucleotide sequences. Fig. 8. Phylogenetic analyses (2421 bases) of rDNA (ITS2 and LSU), partial cp23S and cytochrome b (cob) from S. necroappetens (formerly type A13 or A1.1) distinguish this species lineage from S. microadriaticum (A1). The chloroplast large-subunit (cp23S) further resolved these from other Clade A lineages. The numbers below the branches indicate MP bootstrap values >75. Fig. 9. Genetic differentiation between S. necroappetens and S. microadriaticum as resolved by the psbAncr. The phylogenetic analysis of partial sequences of the PsbA gene (D1 protein of photosystem II) and non-coding region (ncr) of the chloroplast minicircle (region sequenced is indicated by the black colour portion of the graphic that overlays the phylogeny in the upper left-hand corner). PsbAncr haplotypes from each species are resolved into well-separated, highly divergent sequence clusters. Symbols next to each terminal branch correspond to the geographic origin of each S. necroappetens haplotype (seeFig. 5). The numbers below the branches indicate the Bayesian posterior probability (left) and MP bootstrap values (right). Posterior probabilities ≥0.9 are shown.
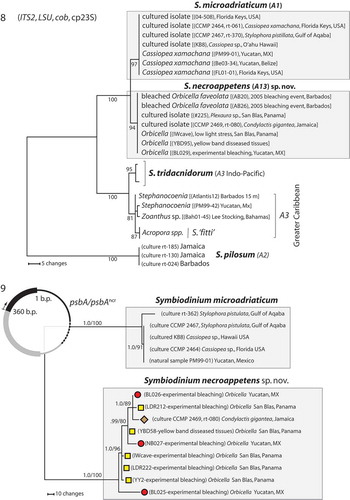
PsbAncr sequence haplotypes of S. necroappetens differed considerably and aligned poorly with the haplotypes of S. microadriaticum (, GenBank accession nos KP165606–KP165619). The sequence variation among haplotypes of S. necroappetens indicated that genotypic diversity was potentially high within this species. No phylogeographic pattern was emergent, which suggested that populations of this species were well connected. In summary, the phylogenetic concordance between genetic markers (rDNA, cob, PsbAncr) designated S. necroappetens as reproductively isolated from S. microadriaticum.
Morphology of Symbiodinium necroappetens
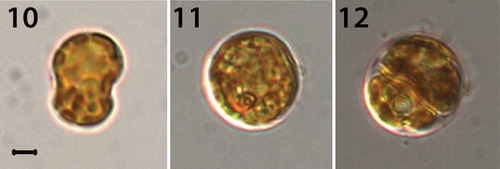
Under an inverted microscope, the motile cells of Symbiodinium necroappetans (strain CCMP2469) showed the typical shape of Symbiodinium spp. Cells were mushroom-shaped when observed ventrally, and had a slightly larger episome (= epicone = epitheca) compared with the hyposome (= hypocone = hypotheca) (–). The cells from a culture in log phase growth (n = 30) were 9.1–12.5 μm in length and 7.5–10.5 μm in width and the length to width ratio of the motile cells was 1.1–1.3 μm (). When fixed and observed using scanning electron microscopy (SEM), the cells were found to be 5.6–12.6 μm in length and 4.3–9.2 μm in width and the length to width ratio was 1.1–1.4 (; –). The elongated amphiesmal vesicle (EAV) located on the apical plate was bordered by three apical plates (the 2′4′ plates), and small x-plate associated in the ventral part (, , ). The 2′ and 4′ plates each possess a row of small knob-like protuberances (). The size of the EAV measured under SEM was 0.89–2.75 μm in length and 0.14–0.38 μm in width, and numbers of knobs were 7–12 ().
Figs 10–12. Light micrographs of S. necroappetens CCMP 2469 (rt-080). Fig. 10. A mastigote (motile) stage. Fig. 11. A coccoid (spherical) stage. Fig. 12. A doublet (dividing) cell. These cells contain a reticulated chloroplast located at the periphery of the cell. Scale bar = 1 μm.
Figs 13–20. Scanning electron micrographs of the motile cells of strain CCMP 2469. Fig. 13. Ventral view showing plate patterns on the episome, cingulum, sulcus and hyposome. Fig. 14. Ventral-left lateral view showing the episome, cingulum, sulcus and hyposome. Fig. 15. Dorsal view showing the episome, cingulum and hyposome. Fig. 16. Ventral-right view. Fig. 17. Apical view showing the episome and elongated amphiesmal vesicle (EAV) plate. Fig. 18. Apical view showing the EAV plate with rows of small knobs (arrowheads) on the 4′ and 2′ plates that align in parallel with the knobs of the EAV. Fig. 19. Antapical view showing plat arrangement on the hyposome. Fig. 20. Antapical-ventral view showing magnified view of the sulcus and peduncle (Pe). The transverse (Tf) and longitudinal (Lf) flagella are labelled on some images. All scale bars = 1 μm.
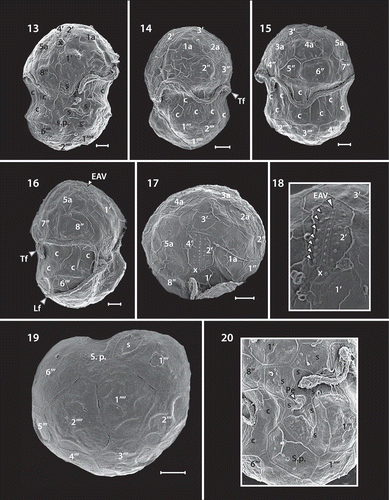
Table 1. Morphological comparison of Clade A Symbiodinium species based on figures obtained under SEM analyses.
The Kofoidian plate formula of S. necroappetens cells was x-plate, EAV, 4′, 5a, 8′′, 9–11s, 21c, 6′′′ and 2′′′′ (, –). The EAV located in the apex of cell, possessed a line of 7–12 knobs in the middle with two more lines located in each side of EAV (, ). The EAV was 0.89–2.75 μm in length and 0.14–0.38 μm in width (). The EAV was bordered by the x-plate and the apical amphiesmal plates 2′ and 4′ plates (, , ). In the episome four apical plates, five intercalary plates and eight precingular plates were present. In the apical plates, a rhomboid shaped 1′, pentagonal shaped 2′, hexagonal shaped 3′ and quadrangular shaped 4’ were located (, ). The intercalary plates were hexagonal (1a, 4a), pentagonal (2a, 3a) and heptagonal (5a) in shape. In the precingular plates, the 1′′ and 4′′ were quadrangular, while 2′′, 3′′, 5′′, 6′′, 7′′ and 8′′ were pentagonal (–, –). Symbiodinium necroappetans possessed about 21 pentagonal shaped cingular plates and 9–11 sulcal plates with peduncle located in the middle of sulcus (–, , –). In SEM micrographs, the cingulum was displaced a 0.06–0.24 proportion of cell length and 0.18–0.72 proportion of cell width (). In the hyposome, six postcingular plates and two antapical plates were present. Most postcingular plates were quadrangular in shape except for 3′′′, which was pentagonal (, ). In the antapical plates, both plates were hexagonal (, ).
Table 2. Morphological analysis of Symbiodinium necroappetens relative to S. microadriaticum and S. pilosum. These data are reprinted from Blank & Huss (Citation1989).
Figs 21–24. Drawings of motile cells showing surface morphology and amphiesmal plate arrangements. Fig. 21. Ventral/sulcal view. Fig. 22. Dorsal view. Fig. 23. Apical view. Fig. 24. Antapical view. Cingulum (c), accessory sulcal plates (s), sulcal or cingulum plates (s?) and posterior sulcal plate (S.p.). All scale bars = 2 µm.
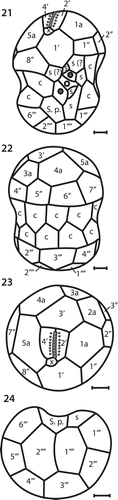
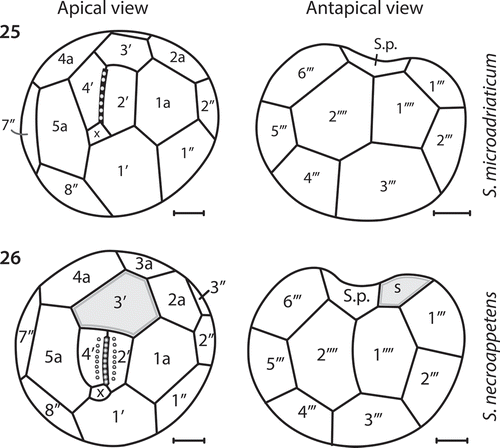
The tabulations for apical, anterior intercalary, precingular, postcingular and antapical plates were identical between S. necroappetens and S. microadriaticum. Their differences in mastigote morphology related to plate shape and arrangement ( and ). Most notably, the size of the anterior intercalary plate, 3′, is considerably larger in S. necroappetens; and on the hyposome (hypotheca), a second sulcal plate contacts the 1′′′′ antapical plate ().
Figs 25–26. Comparison of amphiesmal plate configurations. Fig. 25. The apical and antapical perspectives of S. microadriaticum. Fig. 26. The apical and antapical perspectives of S. necroappetens sp. nov. While the Koifoidian plate formulae are identical between the species (with the possible exception of the cingulum plates), there are differences in plate size and contact points. The enlarged 3′ apical plate of S. necroappetens (highlighted in grey) prevents the 4a anterior intercalary plate from contacting the 4′ apical plate. Two sulcal plates from S. necroappetens contact the 1′′′′ antapical plate, whereas for S. microadriaticum, the posterior sulcal plate alone contacts the antapical plate.
DISCUSSION
The failure to distinguish species diversity has resulted in overly generalized assumptions about the physiology and ecology of Symbiodinium clades. By using the convergence of available genetic, morphological, physiological and ecological evidence, we have described a new species of Symbiodinium whose habit clearly differs from other species within Clade A and expands our understanding of the ecological variation, or niche breadth, in this group.
The recognition of ecological diversity among Symbiodinium
An early reliance on low-resolution genetic markers that resolved only clades of Symbiodinium led to unrealistic generalizations. The merging of ecological attributes from two distinct Clade A species by Toller et al. (Citation2001; who did not resolve the normal mutualistic symbiont, i.e. type A3, from the opportunist, S. necroappetens), when reporting on their experiments with Orbicella, fueled perceptions that members of Clade A were typically stress tolerant, opportunistic, or even parasitic (Stat et al., Citation2008; Lesser et al., Citation2013). The characterization of apparently free-living Clade A species, S. pilosum and S. natans (LaJeunesse, Citation2001; Hansen & Daugbjerg, Citation2009), also suggested that members of the group were less mutualistic, which may have contributed to some of this confusion.
Based on repeated observations, we can begin to reconstruct the ecological habit of S. necroappetens. Several lines of evidence suggest that this species is ecologically cryptic, distributed throughout the Greater Caribbean region living as a non-symbiotic or non-mutualistic opportunist (–). For example, Symbiodinium necroappetens was isolated at independent times from cnidarian tissues during the process of culturing (). The normal (and functional) symbiont that usually resides in these ‘host’ animals either cannot grow in culture or was out-competed by the initially low abundance background cells of S. necroappetens (Santos et al., Citation2001).
When S. necroappetens was reported at detectable densities in nature, it occurred in diseased lesions, especially among colonies of Orbicella spp., or among colonies recovering from severe stress (, ; LaJeunesse et al., Citation2009b; Grotolli et al., 2014). Symbiodinium necroappetens is found in hosts only after cessation of natural or experimental thermal stress, suggesting that conditions must return to normal before it can proliferate. The full or partial necrosis of stressed animal tissues (Brown et al., Citation1995; Hanes & Kempf, Citation2013) generates a high nutrient source that may attract S. necroappetens and allow it to grow opportunistically. Various cultured Symbiodinium exhibit chemo-tropism to nitrogenous sources (Fitt, Citation1984), and therefore many Symbiodinium may exhibit greater mixotrophy without direct access to host inorganic nitrogen. Furthermore, Symbiodinium are capable of feeding phagotrophically on bacteria (Jeong et al., Citation2012). We propose that under natural conditions S. necroappetens may depend on mixotrophy more than ‘symbiotic’ species and that damaged or diseased tissues provide fleeting sources of food either through the uptake of inorganic compounds (D’Elia et al., Citation1983) or by consuming bacteria associated with tissue decomposition. Further observational and experimental evidence is required to adequately assess these possibilities.
The analyses of affected tissues at several Caribbean locations indicate that S. necroappetens is not always present in the necrotic tissues in the yellow band zone, and therefore it is not the causative agent of YBD. It has yet to be detected in the YBD in the Florida Keys (Correa et al., Citation2009; this study). While no trace of this species could be detected in the YBD from Orbicella spp. during a July 2005 sampling in Barbados, it became common among severely bleached and dying colonies in December, 4–6 weeks after the apogee of the 2005 mass bleaching event (LaJeunesse et al., Citation2009b). Symbiodinium necroappetens seems only to have persisted for several months, subsequently being displaced by the proliferation of host-typical Symbiodinium during recovery (LaJeunesse et al., Citation2009b; Grotolli et al., 2014). Therefore, we reason that S. necroappetens exists haphazardly at low abundances in a variety of reef Cnidaria, and probably occurs in the external environment, but proliferates opportunistically under favourable conditions.
ITS rDNA is ‘conserved’ and may poorly resolve species
Some amount of sequence divergence is considered necessary for ribosomal DNA sequence differences to resolve species of dinoflagellate (Litaker et al., Citation2007). Symbiodinium microadriaticum and S. necroappetens are distinguished by differences within the normal intragenomic variation reported for each species (, ), in other Symbiodinium spp. (Thornhill et al., Citation2007; Sampayo et al., Citation2009), and most dinoflagellates (Litaker et al., Citation2007). The significance is that, because the sequences reported here represent the numerically abundant sequence variants comprising a high proportion of rDNA array, they are usually ‘diagnostic’ of evolved species lineages (, ; LaJeunesse & Pinzón, Citation2007). The many remaining sequence variants found in the rDNA array typically consist of functional and non-functional intragenomic variants, as well as sequence artefacts generated by the molecular methods employed to sequence them (Thornhill et al., Citation2007; Arif et al., Citation2014).
Theoretically, most members of a species should share the same dominant rDNA sequences, assuming our understanding about the mechanism of concerted evolution is correct (Dover, Citation1986). However, concerted evolution (under a stochastic model) may conservatively maintain the dominant rDNA sequence in the genomes of individuals that form distinct populations which diverged ecologically and evolutionarily long ago (Leliaert et al., Citation2014). Recent phylogeographic evidence based on independent genetic markers indicate that ITS rDNA can evolve slowly and require many millions of years before nucleotide sequence differences (in the dominant intragenomic variants) arise between two species (Thornhill et al., Citation2014). For example, there are Symbiodinium lineages that share the same diagnostic ITS2 sequence, but appear to have diverged 5–10 million years ago (Thornhill et al., Citation2014). Therefore, the existence of biologically distinct entities with markedly different ecologies (and physiologies) may go unrecognized when thresholds in per cent sequence difference are set arbitrarily for recognizing species (Litaker et al., Citation2007). In the case of S. necroappetens, distinguishing it from S. microadriaticum and type A3 (common to some colonies of Orbicella) dramatically changes our ecological understanding of Clade A species compared with that implied by considering them as a single biological unit (Toller et al., Citation2001; Stat et al., Citation2008; Lesser et al., Citation2013). We now recognize that among the various species that belong to this group, some are free-living (S. pilosum, S. natans), some are opportunistic (S. necroappetens), while others are important mutualistic symbionts (e.g. S. microadriaticum, S. tridacnidorum, including many undescribed species; LaJeunesse et al., Citation2009a).
Comparative morphology, biochemistry and physiology
Using light microscopy, S. necroappetens appears morphologically similar and virtually indistinguishable from many other Symbiodinium in culture. The size range of cells is similar to that of S. microadriaticum (). However, scanning electron microscopy (SEM) identified features that are potentially diagnostic for this species. The ampheismal plate numbers, shapes and configuration were recently reported for Symbiodinium microadriaticum (Lee et al., Citationin press). The Kofoidian plate formula for S. necroappetens is nearly identical to S. microadriaticum, with the apparent exception of having fewer cingulum plates (). There are also clear differences in plate shape and configuration between S. microadriaticum and S. necroappetens that support their distinction as separate species (see , ). The apical plates on S. necroappetens adjoining the EAV (apical plate 2′ and 4′) are adorned with a single row of aligned knobs that appear to differ in morphology from the knobs that run along the axis of the EAV (, ). These features are absent on cells of S. microadriaticum. However, because similar protuberances occur on the apical plates of S. natans (Hansen & Daugbjerg, Citation2009), the taxonomic utility of this feature is questionable. Overall, S. microadriaticum and S. necroappetens are clearly different from S. natans and S. tridacnidorum, supporting their close evolutionary relationship relative to other described Clade A Symbiodinium.
Blank & Huss (Citation1989) first recognized type culture rt-080 (CCMP 2469) as distinct and informally described it as S. microadriaticum subsp. condylactis based primarily on fine-scale differences in cell ultrastructure, number of isoelectric forms of peridinin-chlorophylla-proteins, and the host from which it was cultured. Banaszak et al. (Citation1993) subsequently diagnosed it as a separate species and used the informal designation S. ‘cariborum‘ (nomen nudum). (modified and republished from Blank & Huss, Citation1989) summarizes qualitative and quantitative differences in ultrastructure and protein composition of photosystem pigments between S. necroappetens, S. microadriaticum and the distantly related S. pilosum. Consistent with their genetic similarity, ultrastructural differences between S. necroappetens and S. microadriaticum are equivocal for most character traits and each possesses approximately the same number of chromosomes. The plastid volume of S. necroappetens is slightly less (~18% vs ~22% of total cell volume), but its mitochondrion volume is estimated to be more than triple that of S. microadriaticum and S. pilosum. This indicates that S. necroappetens may be physiologically distinct.
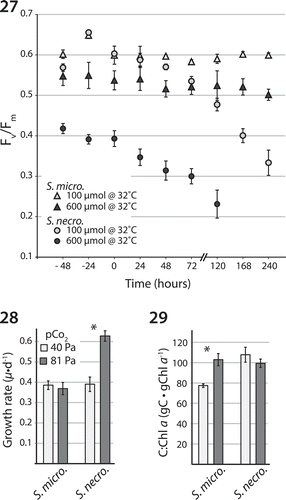
Indeed, significant physiological differences in thermal tolerance, photo-acclimation and response to elevated CO2 are described for S. necroappetens, S. microadriaticum and S. pilosum (Robison & Warner, Citation2006; Hennige et al., Citation2009; Brading et al., Citation2011; Steinke et al., Citation2011). For example, the study of Robison & Warner (Citation2006) assessing photosystem II activity and electron transport, degradation of the D1 protein, and growth, in response to elevated temperature, demonstrated that S. necroappetens was more thermo-sensitive than S. microadriaticum (see also Steinke et al, Citation2011). The thermal tolerance of S. microadriaticum is considerably greater than S. necroappetens under both low (100 µmol photons m−2 s−1) and high (600 µmol photons m−2 s−1) irradiances (; reproduced from Robison & Warner Citation2006), and may relate in part to differences between these species in their ability to acclimate, including the production of antioxidants (Steinke et al., Citation2011). Using both bio-optical and bio-physical analyses, Hennige et al. (Citation2009) compared the responses of S. microadriaticum, S. necroappetens and S. pilosum to changes in photon flux densities. The results are consistent with the interpretation that the three species adjust to changes in the light regime using different mechanisms (see also Iglesias-Prieto & Trench, Citation1994, Citation1997). Finally, Brading et al. (Citation2011) studied the effect of the elevated partial pressure of CO2 (pCO2) on growth and productivity of these species. The steady-state growth rate of S. necroappetens increased by approximately 60% when ambient partial pressure of CO2 was doubled, whereas S. microadriaticum exhibited no significant change in growth rate (; reproduced from Brading et al., Citation2011). However, under this same treatment, the photosynthetic capacity (as assessed by gC•Chla−1) of S. microadriaticum increased significantly while the relative carbon to chlorophyll a of S. necroappetens remained unchanged (). Symbiodinium necroappetens under high pCO2 may be diverting energy away from its carbon concentration mechanisms and reallocating resources for cell division (Brading et al., Citation2011). Assuming that the physiological traits measured for the cultured strains used in these experiments are indicative of each species, they underscore how related Symbiodinium (from the same clade) exhibit large differences in functionality.
Figs 27–29. Physiological trait differences between S. microadriaticum and S. necroappetens sp. nov. Fig. 27. Dark-acclimated PSII quantum yields ± SD (Fv/Fm) measured for each species gradually exposed to elevated temperature and held at 32°C under 100 or 600 µmol photons m−2 s−1 for 10 days (n = 4 technical replicates). Fig. 28. Growth rate (µ) per day under ambient (Pa 40) and (Pa 81) pCO2 atmospheric concentrations projected for the end of this century. Fig. 29. The effect of ambient and high pCO2 on carbon to chlorophyll a concentrations per cell. Asterisk indicates statistically significant differences between treatments for each species. The figures presented here were modified and redrawn from Robison & Warner (Citation2006) and Brading et al. (Citation2011).
No potential conflict of interest was reported by the author(s).
AUTHOR CONTRIBUTIONSM
TCL and DLG made the initial discovery. NK provided key samples from Panama to confirm discovery. SYL and HJJ performed morphological characterizations and TCL conducted molecular analyses. Finally TCL drafted paper and coordinated manuscript revisions with his co-authors.
ACKNOWLEDGEMENTS
We thank Robert K. Trench for thoughtful comments on earlier drafts of the manuscript. Rudolf Blank helped with formulating the species name. Michael McGinley and Mark Warner (U. Delaware) provided samples from Puerto Morelos, MX; Robin Smith collected samples of YBD from the Florida Keys; Ernesto Weil and Garriet W. Smith helped with YBD collections in Puerto Rico; Hazel Oxenford and Jennifer Finney made collections in Barbados during and following the 2005 Caribbean mass coral bleaching event. Mary Alice Coffroth (SUNY Buffalo) kindly provided cultures of S. necroappetens from Panama.
Additional information
Funding
REFERENCES
- Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C.J., LaJeunesse, T.C. & Voolstra, C.R. (2014). Assessing Symbiodinium diversity in scleractinian corals via next generation sequencing-based genotyping of the ITS2 rDNA region. Molecular Ecology, 23: 4418–4433.
- Baker, A.C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annual Review of Ecology, Evolution, and Systematics, 661–689.
- Banaszak, A.T., Iglesias-Prieto, R. & Trench, R.K. (1993). Scrippsiella velellae sp. nov. (Peridiniales) and Gloeodinium viscum sp. nov. (Phytodiniales), dinoflagellate symbionts of two hydrozoans (Cnidaria). Journal of Phycology, 29: 517–528.
- Baums, I.B., Devlin‐Durante, M.K., & LaJeunesse, T.C. (2014). New insights into the dynamics between reef corals and their associated dinoflagellate endosymbionts from population genetic studies. Molecular Ecology 23: 4203–4215.
- Blank, R. J. (1987). Presumed gametes of Symbiodinium: feintings by a fungal parasite? Endocytobiosis and Cell Research, 4: 297–304
- Blank, R.J. & Huss, V.A.R. (1989). DNA divergency and speciation in Symbiodinium (Dinophyceae). Plant Systems Evolution, 163: 213–232.
- Brading, P., Warner, M.E., Davey, P., Smith, D.J., Achterberg, E.P. & Sugget D.J. (2011). Differential effects of ocean acidification on growth and photosynthesis among phylotypes of Symbiodinium (Dinophyceae). Limnology and Oceanography, 56: 927–938.
- Brown, B.E., LeTissier, M.D.A. & Bythell, J.C. (1995). Mechanisms of bleaching deduced from histological studies of reef corals sampled during a natural bleaching event. Marine Biology, 122: 655–663.
- Correa, A.M.S., Brandt, M.E., Smith, T.B., Thornhill, D.J. & Baker, A.C. (2009). Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs, 28: 437–448.
- D’Elia, C.F., Domotor, S.L. & Webb, K.L. (1983). Nutrient uptake kinetics of freshly isolated zooxanthellae. Marine Biology, 75: 157–167.
- Dover, G.A. (1986) Molecular drive in multigene families: how biological novelties arise, spread and are assimilated. Trends in Genetics, 2: 159–165.
- Finney, J.C., Pettay, T., Sampayo, E.M., Warner, M.E., Oxenford, H. & LaJeunesse, T.C. (2010). The relative significance of host-habitat, depth, and geography on the ecology, endemism and speciation of coral endosymbionts. Microbial Ecology, 60: 250–263.
- Fitt, W.K. (1984). The role of chemosensory behavior of Symbiodinium microadriaticum intermediate hosts, and host behavior in the infection of coelenterates and molluscs with zooxanthellae. Marine Biology, 81: 9–17.
- Grottoli, A.G., Warner, M.E., Levas, S.J., Aschaffenburg, M.D, Schoepf, V., McGinley, M., Baumann, J. & Matsui, Y. (2014). The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Global Change Biology, 20: 3823–3833.
- Hanes, S.D. & Kempf, S.C. (2013). Host autophagic degradation and associated symbiont loss in response to heat stress in the symbiotic anemone, Aiptasia pallida. Invertebrate Biology, 132: 95–107.
- Hansen, G. & Daugbjerg, N. (2009). Symbiodinium natans sp. nov.: a free-living dinoflagellate from Tenerife (northeast-Atlantic Ocean). Journal of Phycology, 45: 251–263.
- Hennige, S.J., Suggett, D.J., Warner, M.E., McDougall, K.E. & Smith, D.J. (2009). Photobiology of Symbiodinium revisited: bio-physical and bio-optical signatures. Coral Reefs, 28: 179–195.
- Hirose, M., Reimer, J.D., Hidaka, M. & Suda, S. (2008). Phylogenetic analyses of potentially free-living Symbiodinium spp. isolated from coral reef sand in Okinawa, Japan. Marine Biology, 155: 105–112.
- Huelsenbeck, J.P. & Ronquist, F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics, 17: 754–755.
- Iglesias-Prieto, R. & Trench, R.K. (1994). Acclimation and adaptation to irradiance in symbiotic dinoflagellates. I. Responses of the photosynthetic unit to changes in photon flux density. Marine Ecology Progress Series, 113: 163–175.
- Iglesias-Prieto, R. & Trench, R.K. (1997). Acclimation and adaptation to irradiance in symbiotic dinoflagellates. II. Response of chlorophyll-protein complexes to different photon-flux densities. Marine Biology, 130: 23–33.
- Jeong, H.J., Yoo, Y.D., Kang, N.S., Lim, A.S., Seong, K.A., Lee, S.Y., Lee, M.J., Lee, K.H., Kim, H.S., Shin, W.G., Nam, S.W., Yih, W.H. & Lee, K. (2012). Heterotrophic feeding as a newly identified survival strategy of the dinoflagellate Symbiodinium. Proceedings of the National Academy of Sciences USA, 109: 12604–12609.
- Jeong, H.J., Lee, S.Y., Kang, N.S., Yoo, Y.D., Lim, A.S., Lee, M.J., Kim, H.S., Yih, W.H. & LaJeunesse, T.C. (2014). Genetics and morphology characterize the dinoflagellate Symbiodinium voratum, n. sp., (Dinophyceae) as the sole representative of Symbiodinium clade E. Journal of Eukaryotic Microbiology, 61: 75–94.
- Kang, N.S., Jeong, H.J., Moestrup, Ø., Shin, W.G., Nam, S.W., Park, J.Y., de Salas, M.F., Kim, K.W. & Noh, J.H. (2010). Description of a new planktonic mixotrophic dinoflagellate Paragymnodinium shiwhaense n. gen., n. sp. from the coastal waters off western Korea: morphology, pigments, and ribosomal DNA gene sequence. Journal of Eukaryotic Microbiology, 57: 121–144.
- Kemp, D.W., Fitt, W.K. & Schmidt, G.W. (2008). A microsampling method for genotyping coral symbionts. Coral Reefs, 27: 289–293.
- LaJeunesse, T.C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the internal transcribed spacer region: in search of a ′′species′′ level marker. Journal of Phycology, 37: 866–880.
- LaJeunesse, T.C. (2002). Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Marine Biology, 141: 387–400.
- LaJeunesse T.C. & Pinzón, J.H. (2007). Screening intragenomic rDNA for dominant variants can provide a consistent retrieval of evolutionarily persistent ITS (rDNA) sequences. Molecular Phylogenetics and Evolution, 45: 417–422.
- LaJeunesse, T.C. & Thornhill, D.J. (2011). Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS ONE, 6: e29013.
- LaJeunesse, T.C. & Trench, R.K. (2000). The biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal anemone, Anthopleura elegantissima (Brandt). Biological Bulletin, 199: 126–134.
- LaJeunesse, T.C., Loh, W.K.W., van Woesik, R., Hoegh-Guldberg, O., Schmidt, G.W. & Fitt, W.K. (2003). Low symbiont diversity in southern Great Barrier Reef corals relative to those of the Caribbean. Limnology and Oceanography, 48: 2046–2054.
- LaJeunesse, T.C., Loh, W. & Trench, R.K. (2009a). Do introduced endosymbiotic dinoflagellates ‘take’ to new hosts? Biological Invasions, 11: 995–1003.
- LaJeunesse, T.C., Smith, R., Finney, J.C. & Oxenford, H. (2009b). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proceedings of the Royal Society B, 276: 4139–4148.
- LaJeunesse T. C., Pettay, T., Sampayo, E. M., Phongsuwan, N., Brown, B., Obura, D., Hoegh-Guldberg, O., & Fitt, W. K. (2010). Long-standing environmental conditions, geographic isolation and host–symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr., 37: 785–800.
- LaJeunesse, T.C., Parkinson, J.E. & Reimer, J.D. 2012. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with Cnidaria. Journal of Phycology, 48: 1380–1391.
- LaJeunesse, T. C., Wham, D. C., Pettay, D. T., Parkinson, J. E., Keshavmurthy, S., Chen, C. A. (2014) Ecologically differentiated stress tolerant endosymbionts in the dinoflagellate genus Symbiodinium Clade D are different species. Phycologia 53: 305–319.
- Lee, S.Y., Jeong, H.J., Kang, N.S., Jang, T.Y., Jang, S.H. & LaJeunesse, T.C. (2015). Symbiodinium tridacnidorum sp. nov., a dinoflagellate common to Indo-Pacific giant clams, and a revised morphological description of Symbiodinium microadriaticum Freudenthal, emended Trench et Blank. European Journal of Phycology, doi:10.1080/09670262.2015.1018336.
- Leliaert, F., Verbruggen, H., Vanormelingen, P., Steen, F., Lopez-Bautista, J.M., Zuccarello, G.C. & DeClerk, O. (2014). DNA-based species delimitation in algae. European Journal of Phycology, 49: 179–196.
- Lesser, M.P., Stat, M. & Gates, R.D. (2013). The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs, 32: 603–611.
- Litaker, R.W., Vandersea, M.W., Kibler, S.R., Reece, K.S., Stokes, N.A., Lutzoni, F.M. Yonish, B.A., West, M.A., Black, M.N.D. & Tester, P.A. (2007). Recognizing dinoflagellates species using ITS rDNA sequences. Journal of Phycology, 43: 344–355.
- Pochon, X., Stat, M., Takabayashi, M.T., Chasqui, L., Logan, D.D. & Gates, R.D. (2010). Comparison of endosymbiotic and free-living Symbiodinium (Dinophyceae) diversity in a Hawaiian reef environment. Journal of Phycology, 46: 53–65.
- Porto, I., Granados, C., Restrepo, J.C. & Sanchez, J.A. (2008). Macroalgal-associated dinoflagellates belonging to the genus Symbiodinium in Caribbean reefs. PLoS ONE, 3: e2160.
- Robison, J.D. & Warner, M.E. (2006). Differential impacts of photoacclimation and thermal stress on the photobiology of four different phylotypes of Symbiodinium (Pyrrhophyta). Journal of Phycology, 42: 568–579.
- Rowan, R. & Powers, D. A. (1991). A molecular genetic classification of zooxanthellae and the evolution of animal-algal symbiosis. Science, 251: 1348–1351.
- Rowan, R., Knowlton, N., Baker, A. & Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature, 388: 265–269.
- Santos, S.R., Taylor, D.J. & Coffroth, M.A. (2001). Genetic comparisons of freshly isolated vs. cultured symbiotic dinoflagellates: implications for extrapolating to the intact symbiosis. Journal of Phycology, 37: 900–912.
- Sampayo, E., Dove, S. & LaJeunesse, T.C. (2009). Cohesive molecular genetic data delineate species diversity in the dinoflagellate genus Symbiodinium. Molecular Ecology, 18: 500–519.
- Stat, M., Morris, E. & Gates, R. D. (2008). Functional diversity in coral–dinoflagellate symbiosis. Proceedings of the National Academy of Sciences, 105: 9256–9261.
- Stat, M., Baker, A.C., Bourne, D.G., Correa, A. M., Forsman, Z., Huggett, M.J., Pochon, X., Skillings, D., Toonen, R.J., van Oppen, M.J.H. & Gates, R.D. (2012). Molecular Delineation of Species in the Coral Holobiont. Advances in Marine Biology, 63:1-65.
- Steinke, M., Brading, P., Kerrison, P., Warner, M.E. & Suggett, D.J. (2011). Concentrations of dimethylsulfoniopropionate and dimethyl sulfide are strain-specific in symbiotic dinoflagellates (Symbiodinium sp., Dinophyceae). Journal of Phycology, 47: 775–783.
- Swofford, D.L. (2002). PAUP* Phylogenetic Analysis Using Parsimony (*and other Methods). Version 4.0a136. Sinauer Associates, Sunderland, MA.
- Thornhill, D.J., LaJeunesse, T.C. & Santos, S.R. (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Molecular Ecology, 16: 5326–5340.
- Thornhill, D.J., Lewis, A., Wham, D.C. & LaJeunesse, T.C. (2014). Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution, 68: 352–367.
- Toller, W.W., Rowan, R. & Knowlton, N. (2001). Repopulation of zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease-associated bleaching. Biological Bulletin, 201: 360–373.
- Trench, R.K. & Blank, R.J. (1987). Symbiodinium microadriaticum Freudenthal, S. goreauii sp. nov., S. kawagutii sp. nov., and S. pilosum sp. nov.: Gymnodinioid dinoflagellate symbionts of marine invertebrates. Journal of Phycology, 23: 469–481.
- Warner, M.E., LaJeunesse, T.C., Robison, J.D. & Thur, R.M. (2006). The ecological distribution and comparative photobiology of symbiotic dinoflagellates from reef corals in Belize: potential implications for coral bleaching. Limnology and Oceanography, 51: 1887–1897.
- Xiang, T., Hambleton, E.A., DeNofrio, J.C., Pringle, J.R. & Grossman, A.R. (2013). Isolation of clonal, axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. Journal of Phycology, 49: 447–458.
- Zardoya, R., Costas, E., Lopez-Rodas, V., Garrido-Pertierra, A. & Bautista, J.M. (1995). Revised dinoflagellate phylogeny inferred from molecular analysis of large subunit ribosomal RNA gene sequences. Journal of Molecular Evolution, 41: 637–645.
- Zhang, H., Bhattacharya, D. & Lin, S. (2005). Phylogeny of dinoflagellates based on mitochondrial cytochrome b and nuclear small subunit rDNA sequence comparisons. Journal of Phycology, 41: 411–420.
- Zhang, Z., Green, B.R. & Cavalier-Smith, T. (2000). Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: a possible common origin for sporozoan and dinoflagellate plastids. Journal of Molecular Evolution, 51: 26–41.
