ABSTRACT
Pyropia acanthophora is a foliose Bangiales with widely known endemic populations in Indo-Pacific region. This alga has expanded its range recently as a consequence of introduction. In an attempt to explore the genetic diversity of Py. acanthophora within the Philippines and the impact of the introduction of the species from elsewhere, an examination of molecular differentiation and distribution was undertaken using the mitochondrial COI-5P and plastid rbcL gene sequences. The results revealed that the populations of Py. acanthophora in the Philippines exhibited high haplotypic and genetic diversities, and were found to be distinct from those previously reported as conspecific populations found in Taiwan, India, Japan, Hawaii, and from those introduced populations from Brazil. The network analyses as inferred from rbcL and from the combined COI-5P and rbcL genes showed evidence that the Philippine populations of Py. acanthophora exhibited a chaotic patchiness pattern characterized by a population with highly site-exclusive haplotypes, wide genetic variability and lack of local geographic patterns. The distribution of Py. acanthophora within the Philippines was also found to be greater than what was previously known, ranging from the extreme northwest to extreme northeast mainland Luzon coasts, including Camiguin Is., Cagayan. Understanding the genetic diversity and distribution of Py. acanthophora in the Philippines provides valuable information in relation to the conservation and effective resource management of native populations of Py. acanthophora in the tropical Asian region.
Introduction
The red algal genus Pyropia J. Agardh, Bangiales, includes species of considerable systematic and biogeographic interest due to widespread distribution (Sutherland et al., Citation2011) and economic value of its circumscribed taxa (Hanisak, Citation1998; Mumford & Miura, Citation1998). Notwithstanding the greater species diversity of Pyropia across boreal to cold temperate regions (Brodie et al., Citation1998, Citation2007; Broom et al., Citation1999; Kunimoto et al., Citation1999; Jones et al., Citation2004; Kucera & Saunders, Citation2012; Mols-Mortensen et al., Citation2012, Citation2014), the distribution of the genus extends to some Asian tropical areas, albeit with fewer species (Tanaka & Pham-Hoàng, Citation1962; Cordero, Citation1974; Soe-Htun & Zaw Zaw Pe, Citation1986; Istini et al., Citation1998; Lewmanomont, Citation1998; Ogawa, Citation2001; Kavale et al., Citation2015a, 2015b). The genus is the most speciose of the Bangiales, which currently includes 64 taxonomically accepted species (Guiry & Guiry, Citation2016). The group is notoriously difficult in terms of species discrimination, largely because of wide convergence in almost any available taxonomic diagnostics observed among its members (Sutherland et al., Citation2011). It has been suggested that the total number of taxa or even the number of genera within the Pyropia lineage might be much higher than currently recognized (Sutherland et al., Citation2011).
Molecular genetic data have revealed important insights into the diversity of taxa whose informative phenotypic characters are insufficient for species identification. Especially among members of the genus Pyropia, integrative studies incorporating morphological, molecular and biogeographic data have recently addressed key hypotheses on genetic diversity and distribution (Sutherland et al., Citation2011). Comparative analysis of variable sites of the plastid ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit gene (rbcL) and the 5’ end region of the mitochondrial cytochrome c oxidase I gene (COI-5P) sequences have overcome the limit of morphological identification among the members of Pyropia and have resolved issues on the extent of species distribution (Kucera & Saunders, Citation2012; Guillemin et al., Citation2016).
Although a significant number of Pyropia species are distributed endemically (Yoshida et al., Citation1990; Griffin et al., Citation1999; Kucera & Saunders, Citation2012; Mateo-Cid et al., Citation2012; Nelson & D’Archino, Citation2014; Ramírez et al., Citation2014; Lindstrom et al., Citation2015; Harden et al., Citation2016), there are a few taxa that have been introduced into new areas, which have either not historically been part of their native ranges (Yarish et al., Citation1998; Watson et al., Citation1998, Citation1999; West et al., Citation2005; Verlaque, Citation2001; Neefus et al., Citation2008; Vergés et al., Citation2013a, Citation2013b; Sánchez et al., Citation2014; Nelson et al., Citation2014) or have long been considered as part of their local floras (Brodie et al., Citation1998; Milstein et al., Citation2015). Most of these taxa have been introduced to Atlantic and Mediterranean coasts from the Pacific region and had occurred at an earlier time via global anthropogenic activities () but went unrecorded due to difficulties in species identification (Brodie et al., Citation1998; Neefus et al., Citation2008; Milstein et al., Citation2015). The establishment of the specific sources of the introduced species has also been a challenge due to the lack of information on their corresponding conspecific strains (or identical haplotypes), which represent their assumed native ranges (Neefus et al., Citation2008; Dumilag et al., Citation2016).
Table 1: Summary of the currently known introduced Pyropia species.
Pyropia acanthophora (E. C. Oliveira & Coll) M. C. Oliveira, D. Milstein & E. C. Oliveira was originally described from Ubatuba, São Paulo, Brazil as Porphyra acanthophora (Oliveira Filho & Coll, Citation1975). The current range of distribution of this species has been reported along the warm temperate region of Atlantic Ocean (John et al., Citation2004; Milstein & Oliveira, Citation2005; Creed et al., Citation2010; Sutherland et al., Citation2011; Milstein et al., Citation2012, Citation2015) and Indo-Pacific region (Yoshida, Citation1997, Citation1998; Lee & Kang, Citation2001; Kavale et al., Citation2015a; Sherwood et al., Citation2010; Rivera, Citation2014; Xie et al., Citation2015). For over 40 years, Py. acanthophora was regarded as part of the indigenous flora in Brazil (Oliveira Filho & Coll, Citation1975), but recent molecular analysis has suggested that this species was introduced (Milstein et al., Citation2012, Citation2015). Species identification for Py. acanthophora can be difficult without molecular data (Milstein et al., Citation2015; Xie et al., Citation2015). The presence of Py. acanthophora in the Philippines was recently confirmed using COI-5P gene sequences based on the specimens collected from Cagayan (Rivera, Citation2014). Those specimens may have been described previously as Pyropia denticulata (Levring) J.A. Philips & J.E. Sutherland (Quisumbing, Citation1951) or Pyropia suborbiculata (Kjellman) J.E. Sutherland, H.G. Choi, M.S. Hwang & W.A. Nelson (Cordero, Citation1974; Masuda et al., Citation1991) due to their morphological similarities based on the range of morphology in different habitats, especially on the shape of the thallus and on the presence of a dentate margin of the blade (Levring, Citation1953; Broom et al., Citation2002; Xie et al., Citation2015).
On the basis of phylogenetic analysis, the common ancestry and native lineages of Py. acanthophora are confirmed to occur across much of its known current distributional ranges in the Indo-Pacific region (Milstein et al., Citation2015). While populations of Py. acanthophora from India and Taiwan have been previously examined using COI-5P and rbcL sequences (Kavale et al., Citation2015a; Xie et al., Citation2015), the genetic diversity and distribution of Py. acanthophora within the Philippines are still virtually unknown.
The aim of this study is to describe the genetic differentiation and haplotype distribution of COI-5P and rbcL sequences of Py. acanthophora from the Philippines, including also available sequences of Py. acanthophora from other locations with emphasis on the introduced Py. acanthophora strains.
Materials and methods
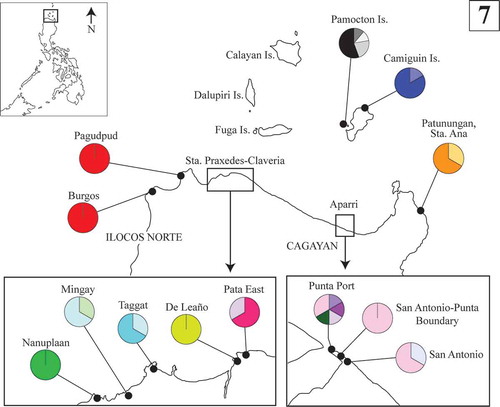
Fifty specimens of Py. acanthophora were collected from 13 sites in the northern Philippines (). Upon collection, a fragment of each specimen was preserved in absolute ethanol in a 1.5 ml microcentrifuge tube. Voucher specimens were prepared following Trono & Ganzon-Fortes (Citation1988) and were deposited in Jose Vera Santos Memorial Herbarium (PUH) with replicates in Far Eastern University Herbarium (FEUH) and in Gregorio T. Velasquez Phycological Herbarium, Marine Science Institute, University of the Philippines, Diliman, Quezon City, Philippines (MSI). Herbarium abbreviations followed Thiers (Citation2016).
Table 2. Summary of sampling locations and haplotype information of Philippine Py. acanthophora specimens used in this study.
DNA extraction was carried out using Qiagen Plant MiniKit (Valencia, CA, USA) following the manufacturer’s protocol. The rbcL primers used in this study included Kito-F1 (Kito et al., the primer sequence published only from GenBank entry AB118578) and JrSR (Broom et al., Citation2010). Four primer combinations were also used to amplify the rbcL gene for samples that did not amplify successfully using the former primers: F67 and R502, F461 and R901, F870 and R1312, and 5rc and rbcspc (Teasdale et al., Citation2002, Vergés et al., Citation2013a); the amplification profiles followed that of Teasdale et al. (Citation2002) and Vergés et al. (2013). For COI-5P gene amplification, the primers GazF1 and GazR1 (Saunders, Citation2005) were used following the profiles used by Hebert et al. (Citation2003). All PCR reactions were prepared in volumes of 30 µl with 1X Vivantis™ Buffer A, 0.1mM dNTPs, 1.5 mM MgCl2 and 0.5 U Taq polymerase. Amplicons were sent to First Base Co. (Selangor, Malaysia) for sequencing. Viewing of chromatograms and contiguous sequence assembly were carried out using DNA Baser version 3.5 (Heracle Biosoft). Generated sequences together with other related sequences from GenBank were aligned using ClustalW (Thompson et al., Citation1994) via the software MEGA ver. 6 (Tamura et al., Citation2011). The COI-5P and rbcL regions of Bangia fuscopurpurea (Dillwyn) Lyngbye isolate GWS001869 (Kucera & Saunders, Citation2012) were used as outgroup sequences for the construction of phylograms. For sequence divergence, percent Kimura-2 parameter (K2P) distances were computed using MEGA ver. 6. The aligned sequences for rbcL (n=70) and COI-5P (n=63) were trimmed to final length of 1317 bp and 642 bp, respectively. A hierarchical-likelihood ratio test was computed in Modeltest 3.04 (Posada, Citation2008) to determine the model of evolution that best fit the sequence data.
Bayesian inference (BI) was estimated using the program MrBayes v3.1.2 (Ronquist & Huelsenbeck, Citation2003). The posterior probabilities were computed using a Metropolis-coupled Markov chain Monte Carlo sampling with 10 million generations. One hundred thousand trees were produced from saving a single tree in every 100 generations. Standard deviation of the split frequency between two simultaneous runs and the Potential Scale Reduction Factor (PSRF) diagnostics was examined to confirm convergence. The first 25% of the trees generated from the burn-in period was discarded and the remaining trees were used to obtain the majority-rule consensus tree. The resulting phylogenetic trees were visualized using FigTree (available http://tree.bio.ed.ac.uk/software).
A statistical parsimony analysis of haplotypes was performed using TCS ver. 1.21 (Clement et al., Citation2000) with a 95% parsimony connection limit and treatment of gaps as missing states. Departure from neutrality on the basis of mismatch distribution (Tajima’s D and Fu & Li’s D* tests) and genetic diversity statistics including haplotype diversity (h), nucleotide diversity (π) and average sequence divergence (k) were computed using DNAsp ver.5 (Librado & Rozas, Citation2009). Haplotype diversity and nucleotide diversity values range from 0 to 1. We considered any population with high h and π diversities if the estimates are greater than 0.5. All figures presented in this study were edited using Adobe® Illustrator® CS5.
Results
Species identification based on interspecific variation and phylogenetic analysis
The intraspecific Kimura-2 parameter (K2P) divergence for the COI-5P of Py. acanthophora ranged from 0–0.7% while its rbcL sequences ranged from 0–0.9%. These divergence ranges were consistent with intraspecific variation reported within Pyropia species (Lindstrom, Citation2008; Kucera & Saunders, Citation2012; Milstein et al., Citation2015 and references therein) and thus the respective sequences could be assigned conspecifically.
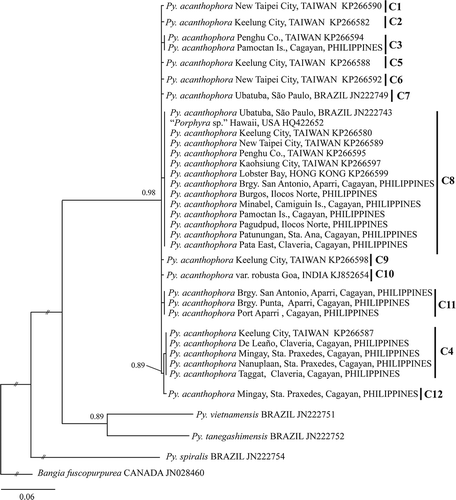
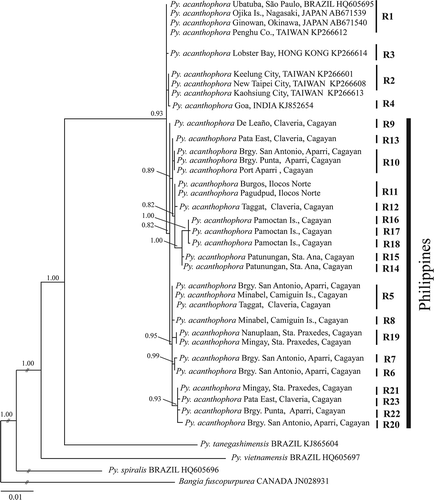
Species identification of our materials was confirmed based on inferred phylogenetic trees using rbcL and COI-5P aligned matrices. The resulting phylogram based on COI-5P dataset showed a distinct lineage for Py. acanthophora with polytomous internodes, representing conspecific samples from the Philippines, Taiwan, Hong Kong, India, Hawaii and Brazil (). All Philippine Py. acanthophora specimens (n = 50) nested within conspecific materials from Brazil, India, Japan, Taiwan and Hong Kong with strong support. Although most of the specimens of Philippine Py. acanthophora formed a clade with those conspecific materials collected from different regions, there were clades that included only Philippine specimens (C11 and C12). The inferred phylogenetic tree based on rbcL dataset showed several lineages for Philippine Py. acanthophora specimens (). Contrary to the result of the COI-5P dataset, the rbcL phylogram indicated that those Philippine Py. acanthophora materials did not group with any of those conspecific specimens collected from other locations.
Haplotype analysis
TCS resolved well-defined networks based on similar datasets used in phylogenetic analysis excluding related Pyropia and outgroup species (Bangia fuscopurpurea). Twelve COI-5P haplotypes (n=84) were defined in a star-like network and lacked apparent geographic clustering. There were five haplotypes observed that represented the Philippine specimens (). The central haplotype C8, found in the introduced Py. acanthophora from Brazil, was detected in 27 samples from the Philippines and from those previously collected materials from Taiwan, Hong Kong and Hawaii. Haplotypes C3 and C4 were found from the Philippines and Taiwan. The haplotype C12 was private, represented by a single specimen from Mingay, Sta. Praxedes, Cagayan, Philippines. Private haplotypes were also recognized from the specimens collected from Brazil (haplotype C7, presumably an introduced strain) and India (haplotype C10).
Figs 3–4. Statistical parsimony analysis representing the entire Py. acanthophora haplotypes inferred from Fig. 3 COI-5P and Fig. 4 rbcL using TCS. The size of the circles represents the number of individuals; black dots correspond to missing haplotypes.
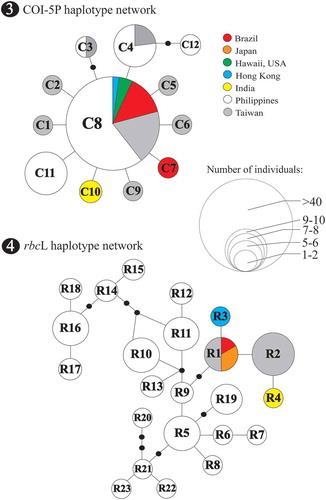
The minimum spanning tree of rbcL haplotypes showed a total number of 23 Py. acanthophora haplotypes from 68 conspecific sequences (). The 50 specimens collected from the Philippines constituted 19 haplotypes with 15 private haplotypes representing site-specific distribution. The introduced Py. acanthophora rbcL haplotype R1 from Brazil was undetected from the Philippine samples but recognized from materials previously sequenced from Japan and Taiwan. One private haplotype each was observed from Taiwan (haplotype R2), Hong Kong (haplotype R3) and India (haplotype R4).
The haplotype network for the combined Py. acanthophora COI-5P-rbcL (referred to herein as combined organellar genes) dataset among endemic populations in Indo-Pacific region revealed 31 distinct sequences including 22 haplotypes for the Philippines, seven for Taiwan and a single haplotype each for Hong Kong and India (). Py. acanthophora populations from India (haplotype I1) and Hong Kong (haplotype H1) were closer to those from Taiwanese Py. acanthophora haplotypes T6 and T1 by a single to two mutation step distances. The Philippine haplotype P1 was separated from Taiwanese haplotype T7 by two mutational steps.
Figs 5–6. Statistical parsimony representing the entire Py. acanthophora haplotypes inferred from the combined COI-5P-rbcL dataset using TCS and their geographic distribution. Fig. 5. Haplotype network for COI-5P-rbcL dataset for all the known native populations of Py. acanthophora. The size of the circles represents the number of individuals; black dots correspond to missing haplotypes. Fig. 6. The geographic distribution of native haplotypes of Py. acanthophora in Hong Kong, Taiwan, and India. In the Philippines, the native populations of Py. acanthophora are limited across Northern Philippines (boxed).
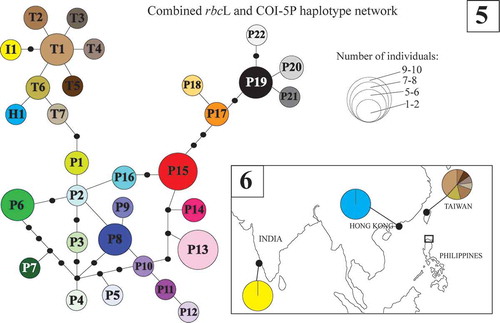
The distribution of haplotypes for the combined organellar gene dataset is shown in and . Although most of the Philippine Py. acanthophora populations appeared specific to particular site, the distribution of private haplotypes within the northern Philippine region was also evident albeit observed only in limited areas. Despite the high number of private haplotypes in the combined organellar genes, no geographic structure within the Philippines was observed.
Genetic differentiation and neutrality test
To analyse genetic diversity of Philippine Py. acanthophora, computation for haplotype diversity (h), nucleotide diversity (π) and average sequence divergence (k) were performed using DNAsp ver.5 (). For all the datasets, the Philippine Py. acanthophora populations showed high haplotype (h) but low nucleotide (π) diversities. The average sequence divergence (k) was relatively higher among the Philippine samples than any other population. As expected, the introduced populations of Py. acanthophora in Brazil represented relatively the lowest h, π and k values. Over all, the rbcL dataset had higher h, π, and k values than the COI-5P dataset. All values computed for Tajima’s D and Fu & Li’s D* neutrality tests were insignificant.
Table 3. Genetic diversity statistics of Py. acanthophora for each geographic location calculated for mitochondrial COI-5P and plastid rbcL sequences.
Discussion
We confirmed a significant increase in the number of known populations and distribution ranges of Py. acanthophora in the Philippines, occurring across northwestern (Ilocos Norte) to northeastern mainland Luzon (Sta. Ana, Cagayan including Pamoctan and Camiguin Islands), based on COI-5P and rbcL sequences. While the COI-5P Py. acanthophora haplotypes from the Philippines were also dominantly found in Taiwan, the rbcL and the combined organellar haplotype network analyses showed clearer discrimination of Philippine Py. acanthophora populations from those conspecific populations from other known geographic areas (e.g. Brazil, Hong Kong, India, Japan and Taiwan).
Surprisingly, the rbcL sequences of Philippine Py. acanthophora specimens showed higher variability than their corresponding COI-5P sequences. Although the common range of rbcL divergence in foliose Bangiales is up to 0.4% (Nelson & Broom, Citation2010; Mols-Mortensen et al., Citation2012; Kucera & Saunders, Citation2012; Lindstrom et al., Citation2015), higher levels such as 0.7% (Guillemin et al., Citation2016) and 1% (Lindstrom, Citation2008) have also been reported. The intraspecific sequence divergences of COI-5P gene in many rhodophytes are commonly higher than rbcL gene (e.g. Geraldino et al., Citation2006; Yang et al., Citation2008; Freshwater et al., Citation2010; Tan et al., Citation2012). However, greater rbcL intraspecies variability (up to 2.1%) might be evidently detected especially on ‘centre of diversity’ sites (McIvor et al., Citation2001). Therefore, the presence of richer genetic diversity among endemic populations of Py. acanthophora in the Philippines, and in Taiwan (Xie et al., 2005) may also suggest that the west Pacific seaboard might be the centre of Py. acanthophora diversity rather than other Indo-Pacific areas.
In all the datasets we used, the haplotype analysis and those computed values that do not significantly differ from zero based on neutrality tests indicated that there exist distinct and demographically stable populations of Py. acanthophora in the Philippines. The greater number of site-specific haplotypes bearing high genetic variability and the absence of geographic patterns observed in Philippine Py. acanthophora populations are typical characteristics of a ‘chaotic patchiness’ pattern (Johnson & Black Citation1982). Although populations that demonstrate chaotic patchiness are more common among marine invertebrates (e.g. Johnson & Black, Citation1982; Arnaud-Haond et al., Citation2008; Muths et al., Citation2009; Fernández et al., Citation2014), some indications of this type of genetic structure especially at small geographic scales, have also been observed for other algal species: Lessonia nigrescens (Martínez et al., Citation2003), Mazaella laminarioides (Faugeron et al., Citation2001) and Portieria hornemanii (Payo et al., Citation2013). The separation of the Philippine populations of Py. acanthophora is likely to be driven by the lack of gene flow prevailing in this area; considering the intricate geological history (Turner et al., Citation2001) and exclusive dynamics of physical factors governing across the northern Philippine coasts (Liang et al., Citation2008; Wang et al., Citation2012; Lu et al., Citation2014).
The presence of identical haplotypes from the northwest Pacific region and Atlantic coasts supports further evidence that Py. acanthophora has been a recent introduction from Pacific seaboards, as previously proposed by Milstein et al. (Citation2015). Analysis of variable sites in COI-5P and rbcL sequences suggested that the introduced populations of Py. acanthophora had lacked genetic variation. The introduced Py. acanthophora strain, which is represented by a single rbcL haplotype is herein recognized as haplotype R1. Similarly, a single cox2-3 intergenic spacer haplotype was also identified for the introduced Py. acanthophora materials from Brazil. However, previous analysis based on the COI-5P haplotype network had recovered two haplotypes (Milstein et al., Citation2012). The very low genetic variation among introduced Py. acanthophora strains suggested that the initial spread was composed of at least two original population sources (haplotypes C7 and C8) from central (i.e. Hawaii) or northwest Pacific (i.e. either from Taiwan or Japan). Although the COI-5P Py. acanthophora haplotype C8 was detected in the Philippines, the corresponding rbcL haplotype assigned for the introduced strain (haplotype R1) was only found in Taiwan and Japan. Whether the original population of the introduced strain came from Hawaii, Taiwan or Japan remains unknown, as the sequenced Py. acanthophora populations from Japan and Hawaii are not well represented. Populations of Py. acanthophora from Korea (Lee & Kang, Citation2001) may shed light to resolving key questions with respect to tracking the original source of the introduced strain. Confirmation on the occurrence of Py. acanthophora from other areas in Atlantic Ocean (e.g. Canary Island, in East Atlantic Ocean: John et al., Citation2004) using molecular datasets may further unravel the extent of the species distribution or possible introduction.
Our findings may contribute towards a deeper understanding on the evolutionary history of tropical foliose Bangiales, especially in the southeast Asian region, where studies on population dynamics of Pyropia species are inadequate. Also, this study may be relevant for planning and designing future development of resource management strategies on this untapped seaweed species of northern Philippines.
Acknowledgements
The authors are grateful to W.D. Monotilla and S.L.Yap for their helpful comments on the manuscript, and to C.B. Mintu, R.D. Olipany, M.P. Quinto, R.C. Andres, and E.C. Ame for their valuable assistance during the sample collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Richard V. Dumilag
R.V. Dumilag: original concept, field work, laboratory experiments, molecular analysis, drafting and editing manuscript; Z.A. Aguinaldo: field work, laboratory experiments, editing manuscript.
Zae-Zae A. Aguinaldo
R.V. Dumilag: original concept, field work, laboratory experiments, molecular analysis, drafting and editing manuscript; Z.A. Aguinaldo: field work, laboratory experiments, editing manuscript.
References
- Aguilar-Rosas, R. & Aguilar-Rosas, L.E. (2003). El género Porphyra (Bangiales, Rhodophyta) en la costa Pacifico di México. I. Porphyra suborbiculata Kjellman. Hydrobiologia, 13: 51–56.
- Arnaud-Haond, S., Vonau, V., Rouxel, C., Bonhomme, F., Prou, J., Goyard, E. & Boudry, P. (2008). Genetic structure at different spatial scales in the pearl oyster (Pinctada margaritifera cumingii) in French Polynesian lagoons: beware of sampling strategy and genetic patchiness. Marine Biology, 155: 147–157.
- Brodie, J., Hayes, P.K., Barker, G.L., Irvine, L.M. & Bartsch, I. (1998). A reappraisal of Porphyra and Bangia (Bangiales, Rhodophyta) in the northeast Atlantic based on the rbcL-rbcS intergenic spacer. Journal of Phycology, 34: 1069–1074.
- Brodie, J., Bartsch, I., Neefus, C., Orfinidis, S., Bray, T. & Mathieson, A. (2007). New insights into the cryptic diversity of the North Atlantic-Mediterranean ‘Porphyra leucosticta’ complex: P. olivii sp. nov. and P. rosengurttii (Bangiales, Rhodophyta). European Journal of Phycology, 42: 3–28.
- Broom, J.R., Jones, W.A., Hill, D.F., Knight, G.A. & Nelson, W.A. (1999). Species recognition in New Zealand Porphyra using 18S rDNA sequencing. Journal of Applied Phycology, 11: 421–428.
- Broom, J.E., Nelson, W.A., Yarish, C., Jones, W.A., Aguilar-Rosas, R. & Aguilar-Rosas, L.E. (2002). A reassessment of the taxonomic status of Porphyra suborbiculata, Porphyra carolinensis and Porphyra lilliputiana (Bangiales, Rhodophyta) based on molecular and morphological data. European Journal of Phycology, 37: 227–235.
- Broom, J.E., Nelson, W.A., Farr, T.J., Phillips, L.E. & Clayton M. (2010). Relationships of the Porphyra (Bangiales, Rhodophyta) flora of the Falkland Islands: a molecular survey using rbcL and nSSU sequence data. Australian Systematic Botany, 23: 27–37.
- Clement, M., Posada, D. & Crandall, K.A. (2000). TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9: 1657–1659.
- Cordero, Jr., P.A. (1974). Phycological observations I: Genus Porphyra of the Philippines, its species and their occurrences. Bulletin of Japanese Society of Phycology, 22: 134–142.
- Creed, M., Fujii, M.T., Barreto, M.B. de B., Guimarães, S.M.P. de B., Cassano, V., Pereira, S.M.B., Carvalho, M. de F., de O. & Khader, S. (2010). Rhodophyceae. In Catálogo de plantas e fungos do Brasil Vol. 1 (Forzza, R.C., editor), 416–436. Andrea Jakobsson Estúdio, Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, Rio de Janeiro.
- Dumilag, R.V., Aguinaldo, Z-Z., Mintu, C.B., Quinto, M.P., Ame, E.C., Andres, R.C., Monotilla, W. D. & Yap, S.L. (2016). Morphological and molecular confirmation of the occurrence of Pyropia tanegashimensis (Bangiales, Rhodophyta) from Palaui Is., Sta. Ana, Cagayan, Philippines. Phytotaxa, 255: 83–90.
- Faugeron, S., Valero, M., Destombe, C., Martínez, E.A. & Correa J.A. (2001). Hierarchical spatial structure and discriminant analysis of genetic diversity in the red alga Mazzaella laminarioides (Gigartinales, Rhodophyta). Journal of Phycology, 37: 705–716.
- Fernández, R., Lemer, S., McIntyre, E. & Giribet, G. (2014). Comparative phylogeography and population genetic structure of three widespread mollusc species in the Mediterranean and near Atlantic. Marine Ecology, 36: 701–715.
- Freshwater, W.D., Tudor, K., O’Shaughnessy, K. & Wysor, B. (2010). DNA barcoding in the red algal order Gelidiales: comparison of COI with rbcL and verification of the “barcoding gap”. Cryptogamie, Algologie, 31: 435–449.
- Geraldino, P.J.L., Yang, E.C. & Boo, S.M. (2006). Morphology and molecular phylogeny of Hypnea flexicaulis (Gigartinales, Rhodophyta) from Korea. Algae, 21: 417–423.
- Griffin, N.J., Bolton, J.J. & Anderson, R.J. (1999). Porphyra aeodis sp. nov. (Bangiales, Rhodophyta), an epiphyte of Aeodes orbitosa from South Africa. European Journal of Phycology, 34: 505–512.
- Guillemin, M., Contreras-Porcia, L. Ramírez, M.E., Macaya, E.C., Contador, C.B., Woods, H., Wyatt, C. & Brodie, J. (2016). The bladed Bangiales (Rhodophyta) of the South Eastern Pacific: molecular species delimitation reveals extensive diversity. Molecular Phylogenetics and Evolution, 94: 814–826.
- Guiry, M.D. & Guiry, G.M. (2016). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 25 May 2016.
- Hanisak, M.D. (1998). Seaweed cultivation: global trends. World Aquaculture Magazine, 29: 18–21.
- Harden, L.K., Morales, K.M. & Hughey, J.R. (2016). Identification of a new marine algal species Pyropia nitida sp. nov. (Bangiales: Rhodophyta) from Monterey, California. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 4: 3058–3062.
- Haroun, R.J., Gil-Rodriguez, M.C., Díaz De Castro J. & Prud’Homme van Reine, W.F. (2002). A checklist of the marine plants from Canary Islands (Central Eastern Atlantic Ocean). Botanica Marina, 45: 139–169.
- Hebert, P.D.N., Cywinska, A., Ball, S.L. & deWaard, J.R. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 270: 313–322.
- Humm, H. J. (1979). The Marine Algae of Virginia, Special Papers in Marine Science No. 3. University of Virginia, Charlottesville.
- Istini, S., Zatnika, A. & Sujatmiko, W. (1998). The seaweed resources of Indonesia. In Seaweed Resources of the World (Critchley, A.T. & Ohno, M., editors), 92–98. Kanagawa International Fisheries Training Centre, Japan International Agency.
- John, D.M., Prud’homme van Reine, W.F., Lawson, G.W., Kostermans, T.B. & Price, J.H. (2004). A taxonomic and geographical catalogue of the seaweeds of the western coast of Africa and adjacent islands. Beihefte zur Nova Hedwigia, 127: 1–339.
- Johnson, M.S. & Black, R. (1982). Chaotic genetic patchiness in an inter-tidal limpet, Siphonaria sp. Marine Biology, 70: 157–164.
- Jones, W.A., Griffin, N.J., Jones, D.T., Nelson, W.A., Farr, T.J. & Broom, J.E. (2004). Phylogenetic diversity in South African Porphyra (Bangiales, Rhodophyta) determined by nuclear SSU sequence analysis. European Journal of Phycology, 39: 197–211.
- Kavale, M.G., Kazi, M.A. & Sreenadhan, N. (2015a). Pyropia acanthophora var. robusta (Bangiales, Rhodophyta) from Goa, India. Indian Journal of Geo-Marine Sciences, 44: 1–8.
- Kavale, M.G., Kazi, M.A., Sreenadhan, N. & Singh, V.V. (2015b). Morphological, ecological and molecular characterization of Pyropia vietnamensis (Bangiales, Rhodophyta) from the Konkan region, India. Phytotaxa, 224: 45–58.
- Kucera, H. & Saunders, G.W. (2012). A survey of Bangiales (Rhodophyta) based on multiple molecular markers reveals cryptic diversity. Journal of Phycology, 48: 869–882.
- Kunimoto, M., Kito, H., Kaminishi, Y., Mizukami, Y. & Murase, N. (1999). Molecular divergence of the SSU rRNA gene and internal transcribed spacer 1 in Porphyra yezoensis (Rhodophyta). Journal of Applied Phycology, 11: 211–216.
- Lee, Y. & Kang, S. (2001). A catalogue of the seaweeds in Korea. Cheju National University Press, Jeju, Korea.
- Levring, T. (1953). The marine algae of Australia. I. Rhodophyta: Goniotrichales, Bangiales and Nemalionales. Arkiv för Botanik, Uppsala, 2: 457–530.
- Lewmanomont, K. (1998). The seaweed resources of Thailand. In Seaweed Resources of the World (Critchley, A.T. & Ohno, M., editors), 70–78. Kanagawa International Fisheries Training Centre, Japan International Agency.
- Liang, W.-D., Yang, Y.J., Tang, T.Y. & Chuang, W.-S. (2008). Kuroshio in the Luzon Strait. Journal of Geophysical Research-Oceans, 113: C08048, 1–19.
- Librado, P. & Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451–1452.
- Lindstrom, S.C. (2008). Cryptic diversity, biogeography and genetic variation in Northeast Pacific species of Porphyra sensu lato (Bangiales, Rhodophyta). Journal of Applied Phycology, 20: 951–962.
- Lindstrom, S.C., Hughey, J.R. & Aguilar Rosas, L.E. (2015). Four new species of Pyropia (Bangiales, Rhodophyta) from the west coasts of North America: the Pyropia lanceolata species complex updated. PhytoKeys, 52: 1–22.
- Lu, W., Yan, X.-H. & Jiang, Y. (2014). Winter bloom and associated upwelling northwest of the Luzon Island: a coupled physical-biological modeling approach. Journal of Geophysical Research-Oceans, 120: 533–546.
- Martínez, E.A., Cárdenas, L. & Pinto, R. (2003). Recovery and genetic diversity of the intertidal kelp Lessonia nigrescens (Phaeophyceae) 20 years after El Niño 1982/83. Journal of Phycology, 39: 504–508
- Masuda, M., Ohno, M. & Trono, G.C. Jr. (1991). A taxonomic assessment of Porphyra suborbiculata Kjellman, a food species from the Philippines. Japan Journal of Phycology, 39: 375–380.
- Mateo-Cid, L.E., Mendoza-Gonzalez, A.C., Díaz-Larrea, J., Sentíes, A., Pedroche, P.F. & Sánchez Heredia, J.D. (2012). A new species of Pyropia (Rhodophyta, Bangiaceae), from the Pacific coast of Mexico, based on morphological and molecular evidence. Phytotaxa, 54: 1–12.
- McIvor, L., Maggs, C.A., Provan, J. & Stanhope, M.J. (2001). rbcL sequences reveal multiple cryptic introductions of the Japanese red alga Polysiphonia harveyi. Molecular Ecology, 10: 911–919.
- Milstein, D. & Oliveira, M.C. (2005). Molecular phylogeny of Bangiales (Rhodophyta) based on small subunit rDNA sequencing: emphasis on Brazilian Porphyra species. Phycologia, 44: 212–221.
- Milstein, D., Medeiros, A., Oliveira, E.C. & Oliveira, M.C. (2012). Will a DNA barcoding approach be useful to identify Porphyra species (Bangiales, Rhodophyta)? A case study with Brazilian taxa. Journal of Applied Phycology, 24: 837–845.
- Milstein, D., Madeiros, A.S., Oliveira, E.C. & Oliveira, M.C. (2015). Native or introduced? A re-evaluation of Pyropia species (Bangiales, Rhodophyta) from Brazil based on molecular analysis. European Journal of Phycology, 50: 37–45.
- Mols-Mortensen A., Neefus, C.D., Nielsen, R., Gunnarsson K., Egilsdóttir, Pedersen P.M. & Brodie J. (2012). New insights into the biodiversity and genetic relationships of foliose Bangiales (Rhodophyta) in Iceland and the Faroe Islands. European Journal of Phycology, 47: 146–159.
- Mols-Mortensen A., Neefus, C.D., Pedersen P.M. & Brodie, J. (2014). Diversity and distribution of foliose Bangiales (Rhodophyta) in West Greenland: a link between North Atlantic and North Pacific. European Journal of Phycology, 49: 1–10.
- Mumford, T.F. Jr. & Miura, A. (1998). Porphyra as food: cultivation and economics. In Algae and Human Affairs (Lembi, C.A. & Waaland, J.R., editors), 87–117. Cambridge University Press, Cambridge.
- Muths, D., Jollivet, D., Gentil, F. & Davoult, D. (2009). Large-scale genetic patchiness among NE Atlantic populations of the brittle star Ophiothrix fragilis. Aquatic Biology, 5: 117–132.
- Neefus, C.D., Mathieson, A.C., Bray, T.L. & Yarish, C. (2008). The distribution, morphology, and ecology of three introduced Asiatic Porphyra (Bangiales, Rhodophyta) in the northwestern Atlantic. Journal of Phycology, 44: 1399–1414.
- Nelson, W.A. & Broom, J.E.S. (2010). The identity of Porphyra columbina (Bangiales, Rhodophyta) originally described from the New Zealand subantarctic islands. Australian Systematic Botany, 23: 16–26.
- Nelson, W.A. & D’Archino, R. (2014). Three new macroalgae from the Three Kings Islands New Zealand including the first southern Pacific Ocean record of the Furcellariaceae (Rhodophyta). Phycologia, 53: 602–613.
- Nelson, W.A., Sutherland, J.E., Hwang, M.S. & Choi, H.-G. (2014). New distributional record for Pyropia koreana: confirmed to occur on the South Island, New Zealand. Algae, 29: 177–181.
- Ogawa, H. (2001). Porphyra in South Asian Waters. In Proceedings of the 11th JSPS Joint Seminar on Marine Science (Taira, M.A., Uematsu, M., Michida, Y. & Kaneko, T., editors), 94–101. Center for International Cooperation, Ocean Research Institute, University of Tokyo, Tokyo, Japan.
- Oliveira Filho, E.C. de & Coll, J. (1975). The genus Porphyra C. Ag. (Rhodophyta, Bangiales) in the American south Atlantic I. Brazilian species. Botanica Marina, 18: 191–197.
- Payo, D.A., Leliaert, F., Verbruggen, H., D’hondt, S. & Calumpong, H.P. (2013). Extensive cryptic species diversity and fine-scale endemism in the marine red alga Portieria in the Philippines. Proceedings of the Royal Society B, 280: 20122660.
- Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25: 1253–1256.
- Quisumbing, E. (1951). Medicinal plants of the Philippines. Republic of the Philippines, Department of Agriculture and Natural Resources, Technical Bulletin, 16: 1–1234.
- Ramírez, M.E., Contreras Porcia, L., Guillemin, M.-L., Brodie, J., Valdivia, C., Flores Molina, M.R., Núñez, A., Bulboa Contador, C. & Lovazzano, C. (2014). Pyropia orbicularis sp. nov. (Rhodophyta, Bangiaceae) based on a population previously known as Porphyra columbina from the central coast of Chile. Phytotaxa, 158: 133–153.
- Rivera, R.A. (2014). Physicochemical properties and quality evaluation of Porphyra acanthophora in the Philippines. MSc. thesis. Pukyong National University.
- Ronquist, F. & Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572–1574.
- Sánchez, N., Vergès, A., Peteiro, C., Sutherland, J. & Brodie, J. (2014). Diversity of bladed Bangiales (Rhodophyta) in western Mediterranean: recognition of the genus Themis and descriptions of T. ballesterosii sp. nov., T. iberica sp. nov., and Pyropia parva sp. nov. Journal of Phycology, 50: 908–929.
- Saunders, G.W. (2005). Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society London Biology, 360: 1879–1888.
- Sherwood, A.R., Kurihara, A., Conklin, K.Y., Sauvage, T. & Presting, G.G. (2010). The Hawaiian Rhodophyta biodiversity survey (2006–2010): a summary of principal findings. BMC Plant Biology, 10: 258.
- Soe-Htun, U. & Zaw Zaw Pe, U. (1986). Studies on the cultivation of conchocelis filaments in Porphyra crispata Kjellman. Burma Research Congress.
- Sutherland, J.E., Lindstrom, S.C., Nelson, W.A., Brodie, J., Lynch, M.D.J., Hwang, M.S., Choi, H., Miyata, M., Kikuchi, N., Oliveira, M.C., Farr, T., Neefus, C., Mols-Mortensen, A., Milstein, D. & Müller, K.M. (2011). A new look at an ancient order: generic revision of the Bangiales (Rhodophyta). Journal of Phycology, 47: 1131–1151.
- Tamura, K., Peterson. D., Peterson. N., Stecher, G., Nei. M. & Kumar. S. (2011). MEGA.5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28: 2731–2739.
- Tan, J., Lim, P.-E., Phang, S.-M., Hong, D.D., Sunarpi, H. & Hurtado, A.Q. (2012). Assessment of four molecular markers as potential DNA barcodes for red algae Kappaphycus Doty and Eucheuma J. Agardh (Solieriaceae, Rhodophyta). PLoS ONE, 7: e52905.
- Tanaka, T. & Pham-Hoàng, H. (1962). Notes on some marine algae from Viet-Nam-I. Memoirs of the Faculty of Fisheries, Kagoshima University, 11: 24–40.
- Teasdale, B., West, A., Taylor, H. & Klein, A. (2002). A simple restriction fragment length polymorphism (RFLP) assay to discriminate common Porphyra (Bangiophyceae, Rhodophyta) taxa from Northwest Atlantic. Journal of Applied Phycology, 14: 293–298.
- Thiers, B. (2016). Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/.
- Thompson, J.D., Higgins, D.J. & Gibson, T.J. (1994). CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research, 22: 4673–4680.
- Torrano-Silva, B.N., Amancio, C.E. & Oliviera, E.C. (2010). Exotic marine macroalgae on Brazilian coast: a revision. Oecologia Australis, 14: 403–414.
- Trono, G.C. Jr. & Ganzon-Fortes, E.T. (1988). Philippine Seaweeds. National Book Store, Manila.
- Turner, H., Hovenkamp, P. & van Welzen, P.C. (2001). Biogeography of Southeast Asia and the West Pacific. Journal of Biogeography, 28: 217–230.
- Vergés, A., Sánchez, N., Peteiro, C., Polo, L. & Brodie, J. (2013a). Pyropia suborbiculata (Bangiales, Rhodophyta): first records from the northeastern Atlantic and Mediterranean of this North Pacific species. Phycologia, 52: 121–129.
- Vergés, A., Comalada, N., Sánchez, N. & Brodie, J. (2013b). A reassessment of the foliose Bangiales (Rhodophyta) in the Balearic Islands including the proposed synonymy of Pyropia olivii with Pyropia koreana. Botanica Marina, 56: 229–240.
- Verlaque, M. (2001). Checklist of macroalgae of Thau Lagoon (Hérault, France), a hot spot of marine species introduction in Europe. Oceanologica Acta, 24: 291–249.
- Wang, G., Wang, D. & Zhou, T. (2012). Upper layer circulation in the Luzon Strait. Aquatic Ecosystem Health and Management, 15: 39–45.
- Watson, K., Cheney, D. & Levine, I. (1998). Can the aquacultured non-indigenous red alga Porphyra yezoensis recruit in Eastport, Maine? Journal of Phycology, 34 ( Suppl.): 62.
- Watson, K., Levine, I. & Cheney, D. (1999). Biomonitoring of an aquacultured introduced seaweed, Porphyra yezoensis (Rhodophyta, Bangiophycidae) in Cobscook Bay, Maine, USA. In Marine Invasions (Pederson, J., editor), 260–264. Proceedings of the First National Conference, January 24–27, 1999, Massachusetts Institute of Technology Sea Grant College Program, Cambridge, MA.
- West, A.L., Mathieson, A.C., Klein, A.S., Neefus, C.D. & Bray, T.L. (2005). Molecular ecological studies of New England species of Porphyra (Rhodophyta, Bangiales). Beihefte zur Nova Hedwigia, 80: 1–24.
- Wynne, M.J. (2005). A check-list of benthic marine algae of the tropical and subtropical western Atlantic: second revision. Beihefte zur Nova Hedwigia, 129: 1–152.
- Xie, Z., Lin, S., Liu, L., Ang, P.O. Jr & Shyu, J. (2015). Genetic diversity and taxonomy of foliose Bangiales (Rhodophyta) from Taiwan based on rbcL and cox1 sequences. Botanica Marina, 58: 189–2012.
- Yang, E.C., Kim, M.S., Geraldino, P.J., Sahoo, D., Shin, J.-A. & Boo, S.M. (2008). Mitochondrial cox1 and rbcL genes of Gracilaria vermiculophylla (Gracilariaceae, Rhodophyta). Journal of Applied Phycology, 20: 161–168.
- Yarish, C.R., Wilkes, R., Chopin, T., Fei, X.G., Mathieson, A.C., Klein, A.S., Neefus, C.D., Mitman, G.G. & Levine, I. (1998). Domestication of indigenous Porphyra (nori) species for commercial cultivation in Northeastern America. World Aquaculture, 29: 26–29.
- Yoshida, T. (1997). Japanese marine algae: new combinations, new names and new species. Phycological Research, 45: 163–167.
- Yoshida, T. (1998). Marine Algae of Japan. Uchida Rokakuho Publishing Co. Ltd, Tokyo.
- Yoshida, T., Nakajima, Y. & Nakata, Y. (1990). Check-list of marine algae of Japan (revised in 1990). Japanese Journal of Phycology, 38: 269–320.