ABSTRACT
The genus Hariotina was established in 1889 based on the type species Hariotina reticulata. This species was then transferred to Coelastrum by Senn (1899). The genus Hariotina was eventually restored by Hegewald et al. (2002) based on morphological characteristics. Species of Hariotina are ubiquitous in fresh water, especially in some tropical and subtropical areas. The taxonomic position of the genus is now considered to be within the family Scenedesmaceae. However, until now, there was an insufficient understanding of the molecular diversity within this genus, and the related reference sequences in the database were inadequate to resolve the species taxonomy. In this study, 20 strains of Hariotina were identified and successfully cultured in the laboratory. We used a combined approach, joining molecular and morphological analyses, to determine their taxonomic relationships and phylogenetic positions. The molecular analyses based on 18S rDNA+ITS sequences and the tufA gene resolved the analysed strains into at least six species, including the known species H. reticulata and H. polychorda and four new species herein described, i.e. H. hainanensis sp. nov., H. compacta sp. nov., H. guilinensis sp. nov. and H. laxa sp. nov. The genus Hariotina was confirmed in the Scenedesmaceae as a monophyletic clade that was closely related to the genus Coelastrum in tufA gene phylogeny. Strains with H. reticulata morphotype were found to be located on two different branches, and formed sister relationships with the other two morphologically distinct species. In addition, the topology was supported by all three DNA markers. The congruence between the phylogeny and the classical taxonomic system based on morphological criteria for this taxon is discussed.
Introduction
The subfamily Coelastroideae (Scenedesmaceae, Sphaeropleales) was originally characterized by three-dimensionally arranged, more or less spherical coenobia, and it mainly included the genus Coelastrum Nägeli and its relatives (Smith, Citation1920; Printz, Citation1927). Members of this subfamily are widely distributed in freshwater bodies. This assemblage was first treated as a family, Coelastraceae, and a close relationship to the Scenedesmaceae was suggested (Wille, Citation1909). The idea that Coelastrum and its relatives could represent a subfamily in the family Scenedesmaceae was first expressed by Smith (Citation1920) and Printz (Citation1927) based on morphological features. Hegewald et al. (Citation2010) reinstated the subfamily Coelastroideae and included the five genera Coelastrum Nägeli, Coelastrella Chodat, Hariotina Dangeard, Asterarcys Comas and Dimorpho-coccus Braun under this group, based on ITS2 secondary structure phylogeny. Among them, the genus Hariotina was morphologically similar to Coelastrum and it was distinct from the other three genera by its spherical cell shape (different from Dimorphococcus), usual colony (different from Coelastrella) and colony forming by connecting strands (different from Asterarcys).
The relationship between the genera Hariotina and Coelastrum was always considered close (Senn, Citation1899; Hegewald et al., Citation2010). Hariotina was first erected by Dangeard (Citation1889) with the type species H. reticulata P. A.Dangeard. It was then transferred to Coelastrum by Senn (Citation1899). Koršikov (Citation1953) described a variety of that species, Coelastrum reticulatum var. polychordum Koršikov, which differed from the type by more than one connection strand between the cells. Hegewald et al. (Citation2002) restored the genus Hariotina, separating it from Coelastrum based on the position of different connection strands and whether or not there was a colony mucilage layer. In addition, the number of connection strands was considered a sufficient character to distinguish species in Hariotina, and the variety Coelastrum reticulatum var. polychordum was erected to the species level as Hariotina polychorda (Korshikov) E. Hegewald in this genus (Hegewald et al., Citation2002). This was supported by Krienitz et al. (Citation2003) with similar observations. Hegewald et al. (Citation2010) summarized the differences between the genera Hariotina and Coelastrum, which mainly differed in connection strands and mucilage. The genus Hariotina was considered to have one to three thin elongated connection strands on the subapical region of the cells, and the coenobium was embedded in mucilage, whereas Coelastrum had its short wide connection strands at the base of the cells and had no mucilage envelope (Hegewald et al., Citation2010). Now, according to AlgaeBase, Hariotina is considered an accepted genus with two distinct species (Guiry & Guiry, Citation2020).
In this study, 20 strains, sampled from different locations and morphologically ascribable to Hariotina, were collected in China and successfully cultured. The performed phylogenetic and morphological analyses suggested the attribution of the investigated strains to at least six species: H. hainanensis sp. nov., H. compacta sp. nov., H. guilinensis sp. nov., H. laxa sp. nov., H. reticulata and H. polychorda. The aim of this study was to evaluate the molecular diversity of this widespread freshwater planktonic group and discuss the present classification of Hariotina-like taxa by combining molecular and morphological data.
Materials and methods
Microalgal isolation and cultivation
This study was based on 20 samples collected in China. The samples were mainly isolated from fresh water in subtropical and tropical regions in China (). Single cells were isolated from samples using the serial dilution pipetting technique (Hoshaw & Rosowski, Citation1973) under an inverted microscope (CKX41; Olympus, Tokyo, Japan). Cells were maintained in a liquid BG11 medium (Stanier et al., Citation1971) with a constant light source of 30–50 μmol photons m–2 s–1 and a temperature of 25°C. Voucher specimens were fixed in 4% (final concentration) formaldehyde (put in vials) and deposited in the Freshwater Algal Herbarium, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China (HBI); herbarium acronyms follow Thiers (Citation2019). Cultures can be obtained from the Freshwater Algae Culture Collection, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China (FACHB). The voucher numbers are provided in .
Table 1. Location of the collecting sites and GenBank accession number of isolates
DNA extraction, PCR amplification and sequencing
DNA was extracted using an AxyPrep Genomic DNA Miniprep Kit (Axygen, Hangzhou, China). PCR amplification was performed using 3 μl template DNA, 0.1 μM of each primer and 25 μl 2× Tap Master Mix (ExTaq; Takara) in a 50 μl reaction volume. Nuclear-encoded 18S rDNA sequences were amplified using the primers 18F (5′-AACCTGGTTGATCCTGCCAGT-3′) and 18R (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (Medlin et al., Citation1988). The amplification conditions were as follows: 5 min at 94°C, 32 cycles of 50 s at 94°C, 60 s at 55°C, 90 s at 72°C, and a final 10 min extension step at 72°C. The ITS sequence was amplified using the primer pair NS7 m (5′-GGCAATAACAGGTCTGT- 3′) and LR1850 (5′ -CCTCACGGTACTTGTTC- 3′) (Bhattacharya et al., Citation1996). The amplification conditions for ITS were as follows: 5 min at 94°C, 32 cycles of 50 s at 94°C, 60 s at 52°C, 90 s at 72°C, and a final 10 min extension step at 72°C. The tufA sequence was amplified using the primer pair tufAF (5′ -TGAAACAGAAMAWCGTCATTATGC- 3′) and tufAR (5′ -CCTTCNCGAATMGCRAAWCGC- 3′) (Famà et al., Citation2002). The amplification conditions for tufA were as follows: 5 min at 94°C, 32 cycles of 50 s at 94°C, 50 s at 56.5°C, 70 s at 72°C, and a final 10 min extension step at 72°C. PCR products were sequenced by TSINGKE Biotechnologies (China). The sequences were assembled using the ContigExpress program of the Vector NTI software suite (Lu & Moriyama, Citation2004) and deposited in GenBank under the accession numbers provided in .
Morphological observations
An Olympus BX53 (Tokyo, Japan) light microscope with an Olympus DP80 digital camera and CellSens standard image analysis software (Tokyo, Japan) was used for morphological assessment. For scanning electron microscopy (SEM) analysis, samples collected in the habitat were directly fixed with 2.5% glutaraldehyde for 8–10 h, then treated with 2% (wt:vol) solution of cetyltrimethylammonium bromide (CTAB) to remove the mucilage envelope (Tavera & Calderon, Citation2013). Microscope slides (0.5 cm × 0.5 cm) were coated with three layers of poly-L-lysine solution to aid adhesion of the cells. Gradual dehydration of the cells was achieved by dipping them into an ethanol series of 30% (30 min), 50% (30 min), 70% (30 min), 90% (30 min), 95% (30 min), 99% (15 min) and 100% (15 min) concentration (Kaufnerová & Eliáš, Citation2013). Subsequently, the algal cells were critical-point dried with CO2 and sputter-coated with gold. A Hitachi S-4800 (Tokyo, Japan) scanning electron microscope was used to visualize the fixed cells.
Molecular phylogenetic analyses
The nuclear-encoded 18S rDNA+ITS sequences and nucleotide sequence of the tufA gene were used for phylogenetic analyses. Sequences of related species were obtained from GenBank (Supplementary table 1). Members of Hydrodictyaceae were chosen as outgroup. Sequences were aligned using Mafft 7.0 (Katoh & Standley, Citation2013) and adjusted manually using MEGA6 (Tamura et al., Citation2013). Bayesian inference (BI) analyses were estimated using MrBayes version 3.2.1 (Ronquist et al., Citation2012) with the model GTR+I+G suggested by MrModeltest 2.2 (Nylander, Citation2004). Markov Chain Monte Carlo (MCMC) analyses were run with four Markov chains (three heated, one cold) for 1 × 106 generations, with trees sampled every 100 generations. Every time the diagnostics were calculated, a fixed proportion of samples (burninfrac = 0.25) was discarded from the beginning of the chain. Maximum likelihood (ML) trees were inferred via RAxML version 8.0.24 (Stamatakis, Citation2014) with the GTRGAMMAI model. RAxML was implemented with the rapid bootstrapping algorithm (1000 replicates).
For each of the obtained ITS region sequences, the ITS2 boundaries were detected with ITS2 Database (Schultz et al., Citation2006). For each ITS2 sequence, the secondary structure was predicted in ITS2 Database using a consensus model of Coelastrella (Kaufnerová & Eliáš, Citation2013), and was then adjusted manually with VARNA 3.7 (Darty et al., Citation2009). The compensatory base changes (CBCs) and the consensus model of Hariotina were analysed by 4SALE 1.5 (Seibel et al., Citation2006, Citation2008).
Results
Morphological observation and taxonomic implications
Strains of Hariotina described herein are characterized as coenobia of 4–8–16–32–64 cells embedded in a mucilage layer. The cells are spherical or nearly spherical in shape. Adjacent cells are connected apically or subapically by 1–3 narrow connection strands of the outer cell wall layer. Large intercellular spaces are triangular to irregularly rounded in shape. Syncoenobium often develop during reproduction. Based on the morphological investigation combined with phylogenetic analyses, four new species were identified.
Hariotina compacta Wang, Liu, Hu & Liu sp. nov. (–)
Description: Coenobium spherical, with (4)–8–16–(32) cells embedded in a mucilage layer (–). The cells are arranged in a compact arrangement, the coenobium measures 15–40 μm in diameter and the mucilage layer is 2–6 μm thick. Cells spherical, measuring 8–12 μm in diameter. Neighbouring cells are connected with 1–2 subapical strands, which formed during the autosporangium period; the strands are 1.5–2.2 μm in width, and each cell forms 5–8 strands. The connecting strands are arched from the side; the basal part is wider, but a significant constriction occurs at the middle part (the contact site of two strands from different cells) (). Reproduction by forming daughter coenobium of (4)–8–16–(32) cells (). Compound coenobium often develops during reproduction, measuring 55–115 μm in diameter.
Figs 1–4. Morphology of Hariotina compacta. Fig. 1. Sixteen-celled coenobia. Fig. 2. Negative staining with India ink showing mucilage layer. Fig. 3. Eight-celled coenobia, and the arrow indicates the side view of the connection strand. Fig. 4. Syncoenobia. Scale bar = 10 μm
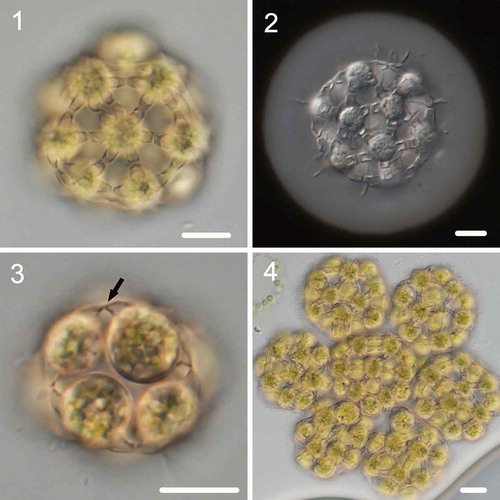
This species differs in morphology from other species of this genus by the wider (> 1.5 μm) connecting strands and more compact cell arrangement. In addition, it has independent phylogenetic positions and forms a sister branch with H. reticulata based on 18S+ITS and tufA gene phylogeny.
Holotype (Here designated): Formaldehyde-fixed specimen BNII5-8, collected from a tropical pond by Qinghua Wang in April 2017, is deposited in the HBI.
Type locality: Jinghong, Yunnan Province N21°55′48″, E101°14′54″.
Habitat: Ponds in the tropical zone.
Etymology: The specific epithet ‘compacta’ (fem. adj.) refers to the compact cell arrangement in morphology.
Reference living culture: FACHB-2353.
Available DNA sequences: FACHB-2353: 18S (MK408681), ITS (MK408701), tufA (MK408721).
Hariotina hainanensis Wang, Liu, Hu & Liu sp. nov. (–, –)
Description: Coenobium spherical, with (4)–8–16–32 cells embedded in a mucilage layer (–). The coenobium measures 15–45 μm in diameter, and the mucilage layer is 2–8 μm thick. Cells spherical, measuring 8–12 μm in diameter. Neighbouring cells are connected with 1–2 subapical strands, which formed during the autosporangium period; the strands are 1–1.3 μm in width, and each cell forms 5–6–(8) strands. The connecting strands are arched from the side; the basal part is wider, but a significant constriction occurs at the middle part (the contact site of two strands from different cells) (). Reproduction by forming daughter coenobium of 4–8–16–32 cells. Compound coenobium often develop during reproduction, measuring 45–80 μm in diameter (). The SEM images are shown in –.
Figs 5–13. Morphology of Hariotina hainanensis and Hariotina reticulata. Figs 5–10. Morphology of Hariotina hainanensis. Fig. 5. Coenobia of strain HND25-3. Fig. 6. Coenobia of strain HND25-3.The arrow indicates the side view of the connection strand. Fig. 7. Coenobia of strain GL1-4. Fig. 8. Coenobia of strain GL1-4. Negative staining with India ink showing mucilage layer. Fig. 9. Coenobia of strain BN9-1. Fig. 10. Autosporangia. Figs 11–13. Morphology of Hariotina reticulata. Figs 11–12. Negative staining with India ink showing mucilage layer. Fig. 13. The side view of the connection strand. Scale bar = 10 μm

This species is very similar to Hariotina reticulata in morphology. However, it has independent phylogenetic positions and forms a sister branch with H. polychorda (morphology of H. polychorda showed in Supplementary fig. 1) based on 18S+ITS and tufA gene phylogeny.
Holotype (Here designated): Formaldehyde-fixed specimen HND25-3, collected from a tropical pond by Qinghua Wang in March 2016, is deposited in the HBI.
Type locality: Danzhou, Hainan Province N19°28′′20″, E109°33′49).
Habitat: Ponds in tropical, subtropical and temperate regions.
Etymology: The species epithet ‘hainanensis’ (fem. adj.) refers to the type locality Hainan Province.
Reference living culture: FACHB-2341.
Available DNA sequences: FACHB-2341: 18S (MK408684), ITS (MK408704), tufA (MK408724).
Hariotina laxa Wang, Liu, Hu & Liu sp. nov. (–, –)
Description: Coenobium spherical or ellipsoidal, hollow inside, with (4)–8–16–32 cells embedded in a mucilage layer (–). The intercellular space is triangular and large, approximately equal to the size of the cells. The coenobium measures 20–45 μm in diameter, and the mucilage layer is 6–15 μm thick. Cells spherical or ellipsoidal from the side, slightly flattened; hexagonal, pentagonal or rounded apex, measuring 7–12 μm in diameter. Neighbouring cells are connected with 1–(2) subapical strands (), which formed during the autosporangium period; the strands are 1.2–2 μm in width, and each cell forms 5–6–(7) strands. The connecting strands are straight or slightly arched from the side, with uniform thickness and no constriction at the contact site (). Reproduction by forming daughter coenobium of (4)–8–16 cells. Compound coenobium often develop during reproduction, measuring 45–110 μm in diameter (). The SEM images of H. laxa are shown in –.
Figs 14–22. Morphology of Hariotina laxa and Hariotina guilinensis. Figs 14–19. Morphology of Hariotina laxa. Fig. 19. Syncoenobia. Figs 20–22. Morphology of Hariotina guilinensis. Fig. 22. Syncoenobia. Scale bar = 10 μm

Figs 23–28. The SEM images of Hariotina laxa and Hariotina hainanensis. Figs 23–25. The SEM images of Hariotina laxa. Figs 23–24. Coenobia of Hariotina laxa (the mucilage layer removed by CTAB). Fig. 23. Hariotina laxa without removing the mucilage layer. Figs 25–28. SEM images of Hariotina hainanensis. Scale bar = 10 μm
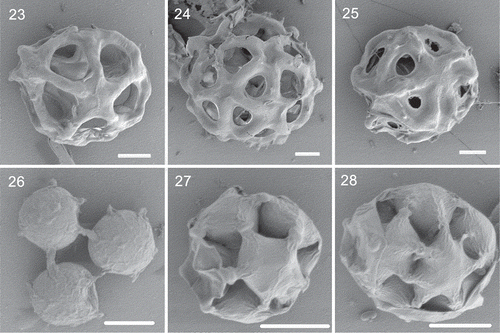
This species differs from other species of this genus by the larger intercellular space and different strand structure in morphology and has independent phylogenetic positions based on 18S+ITS and tufA gene phylogeny.
Holotype (Here designated): Formaldehyde-fixed specimen YY1-2, collected from a subtropical pond by Qinghua Wang in October 2017, is deposited in the HBI.
Type locality: Yueyang, Hunan Province (N29°22′30″, E113°6′36″).
Habitat: Ponds in tropical, subtropical regions.
Etymology: The specific epithet ‘laxa’ (fem. adj.) refers to the loose coenobium structure on morphology.
Reference living culture: FACHB-2346.
Available DNA sequences: FACHB-2346: 18S (MK408692), ITS (MK408712), tufA (MK408732).
Hariotina guilinensis Wang, Liu, Hu & Liu sp. nov. (–)
Description: Coenobium spherical or ellipsoidal, hollow inside, with 4–8–16 cells embedded in a mucilage layer (–). The intercellular space is triangular or quadrangle, slightly smaller than the cell size. The coenobium measures 12–35 μm in diameter, and the mucilage layer is 2–3 μm thick. Cells are subspherical from the side, slightly flattened; pentagonal, hexagonal, tetragonal, rounded and sometimes triangular (seen in 4-celled coenobium) apex, measuring 5–9 μm in diameter. Neighbouring cells are connected with only one lateral strand, which formed during the autosporangium period; the strands are 1–1.2 μm in width, and each cell forms 3–4–5–6 strands. The connecting strands are straight or slightly arched from the side, with uniform thickness and no constriction at the contact site. Reproduction by forming daughter coenobium of 4–8–16 cells. Compound coenobium often develop during reproduction, measuring 40–70 μm in diameter ().
This species shares some morphological features with Hariotina laxa, but it differs from H. laxa by the smaller intercellular space. It also has independent phylogenetic positions based on 18S+ITS and tufA gene phylogeny.
Holotype (Here designated): Formaldehyde-fixed specimen GL28-1, collected from a subtropical fishpond by Qinghua Wang in December 2016, is deposited in the HBI.
Type locality: Guilin, Guangxi Province (N25°24′2″, E110°14′42″).
Habitat: Ponds in tropical, subtropical regions.
Etymology: The species epithet ‘guilinensis’ (fem. adj.) refers to the type locality Guilin, Guangxi Province.
RReference living culture: FACHB-2357.
Available DNA sequences: FACHB-2357: 18S (MK408687), ITS (MK408707), tufA (MK408727).
Molecular phylogeny
The phylogenetic position for each of the strains was inferred by analysing the 18S+ITS sequences and tufA gene sequenceswhich was published earlier. The data sets were analysed using both ML and BI methods, although only the tree resulting from ML analysis is shown. The topologies of trees inferred from ML and BI were congruent, and the ML trees for the 18S+ITS gene and tufA gene are shown in and , respectively, with bootstrap support and posterior probability values indicating branch support.
Fig. 29. Maximum-likelihood phylogenetic tree inferred from 18S+ITS sequences. Only bootstrap values above 50 and Bayesian posterior probabilities above 0.75 are shown. Different colour boxes represent different species in Hariotina. Serial number represents intron insertion type. The strains provided in this study are indicated in bold. Strains labelled with quotes indicate that names correspond to GenBank labels and not confirmed by comparison with type material
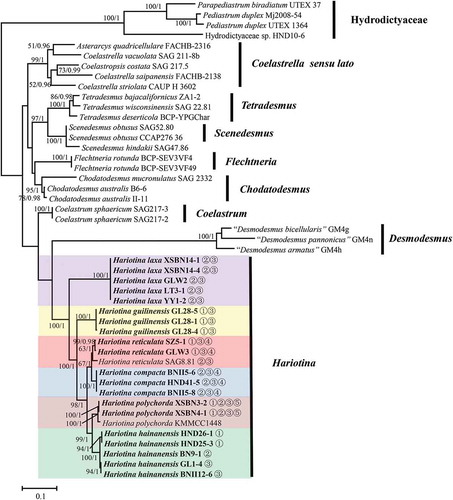
Fig. 30. Maximum-likelihood phylogenetic tree inferred from tufA gene sequences. Only bootstrap values above 50 and Bayesian posterior probabilities above 0.75 are shown. Different colour boxes represent different species in Hariotina. The strains provided in this study are indicated in bold. Strains labelled with quotes indicate that names correspond to GenBank labels and not confirmed by comparison with type material
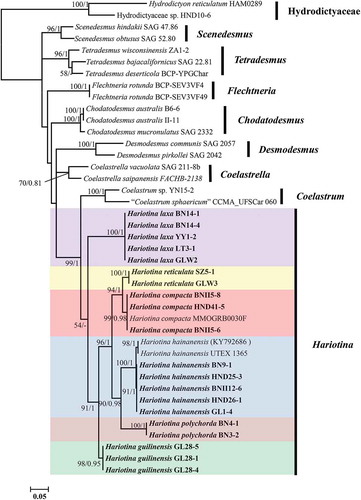
The 18S+ITS phylogenetic tree showed a well-supported (100/1) clade of the genus Hariotina. Both the 18S+ITS rDNA gene and tufA gene phylogenies revealed that there were at least six subclades in the genus Hariotina, which represented six different species. The topology of the two phylogenetic reconstructions within the clade Hariotina was almost identical: within the genus, H. laxa and H. guilinensis were located at the base of the clade, with H. guilinensis more internal; H. reticulata was sister taxon to H. compacta and H. polychorda was sister taxon to H. hainanensis. Considering the analysis based on the 18S rDNA gene alone, H. reticulata could not be distinguished from H. compacta and H. polychorda was not separated from H. hainanensis (Supplementary fig. 2).
Intron insertions in the 18S rDNA gene of this group were very common, with up to four introns inserted in H. polychorda. The presence of introns is indicated by the serial number according to the order in which they are inserted, shown in . The result revealed that there were three ITS genotypes in H. hainanensis, which corresponded to three intron insertion types in the 18S rDNA gene, respectively. However, all their tufA genes were almost identical.
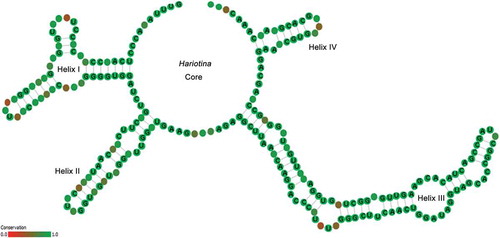
The consensus ITS2 secondary structure model of Hariotina is shown in . The matrix of hemi-compensatory base changes (h-CBCs) and CBCs for different species are presented in Supplementary table 2. Interspecies base changes in Hariotina ranged from 0 to 8 h-CBCs and 0 to 8 CBCs. There were 5–6 h-CBCs and 0–1 CBCs between the sister species H. hainanensis and H. polychorda and they were mainly located in helix I (1–2), helix II (3) and helix III (1), and only 1 h-CBC in helix I between the sister species H. reticulata and H. compacta. In addition, there were only 0–1 h-CBCs in helix I between the three genotypes of H. hainanensis.
Discussion
The genera Hariotina and Coelastrum are common freshwater Coelastroideae taxa found all over the world. For a long time in the past, they were attributed to the genus Coelastrum. Since Hegewald et al. (Citation2002) reintroduced the name Hariotina, this genus has been gradually accepted (Krienitz et al., Citation2003; Hegewald et al., Citation2010). In this study, our findings based on more specimens also support this conclusion, with several Hariotina-like species forming a monophyletic group in Scenedesmaceae, which was separated from the Coelastrum lineage. Species of this genus are mainly distributed in stagnant water in tropical and subtropical regions. Among them, H. compacta and H. hainanensis were also found in temperate regions of northern China. Hariotina polychorda was also found to be widely distributed in large rivers in subtropical areas.
Hariotina reticulata and H. hainanensis are very similar in morphology. It is difficult to distinguish them based solely on morphological characteristics. At present, two nominally H. reticulata strains (SAG 8.81 and UTEX 1365) in the database that have been certified by taxonomists have been proven to be located in different phylogenetic branches. We chose to define the branch containing the strain SAG 8.81, which was published earlier (Hegewald et al., Citation2010), as the type species H. reticulata, and the other branch as H. hainanensis. Although H. hainanensis is more similar to H. reticulata in morphology, the phylogenetic results indicated that it is clustered with H. polychorda, while H. reticulata has a close relationship with H. compacta. That is, both of the above two pairs of sisters (H. reticulata and H. compacta; H. hainanensis and H. polychorda) have obvious morphological differences, while the two species with similar morphology, H. reticulata and H. hainanensis, are distributed in different branches.
In addition to H. reticulata and H. hainanensis, there are another two species that are similar in morphology, H. laxa and H. guilinensis. They differ from other species by the large intercellular space and nearly straight connection strands without constriction. Compared with H. laxa, H. guilinensis has a smaller cell size and intercellular space. However, these characteristics are sometimes susceptible to environmental conditions. Therefore, the molecular data are more reliable for the identification of these two species at present.
The finding that five strains of H. hainanensis contained three ITS genotypes and three intron insertion types in the 18S rDNA gene, but their tufA genes and morphologies are almost identical, has led us to treat these strains as the same species for the present. Furthermore, different ITS genotypes and intron insertion types are only considered as intraspecific variations. The same species possessing multiple intron insertion types has also been reported in many other groups in Chlorophyceae (Friedl et al., Citation2000; Liu et al., Citation2018).
Although there were no CBCs (only h-CBCs) between some closely related species (H. reticulata and H. compacta; H. hainanensis and H. polychorda), they do have stable morphological differences. Taking into account the phylogenetic results and morphological characteristics, we still treat H. compacta and H. hainanensis as distinct species. The phenomenon of two distinct species with no CBCs is common in Scenedesmaceae, such as Coelastrella (Kaufnerová & Eliáš, Citation2013; Wang et al., Citation2019) and Chodatodesmus (Sciuto et al., Citation2015). In addition, the presence and location of CBCs in this taxon are sometimes random. Therefore, the mere presence of CBCs in two strains does not indicate that they represent distinct species, and vice versa. The CBC concept has also been found to be misleading for Ulvophycean taxa (Caisová et al., Citation2011).
So far, the delineation of taxa within the genus Hariotina has been exclusively on the basis of morphological criteria. Here, we provide insight from the molecular diversity of these morphologies to redefine species in this genus. The 18S rDNA gene was not suitable for species detection in this taxon. This conclusion was reached not only because there were multiple intron insertions in this region, which greatly increased the difficulty and cost of sequencing, but also because the 18S rDNA gene has low resolving power at the species level. Now, a joint use of multiple markers seems to be the ideal tool for barcoding cryptic green algal species (Fučíková et al., Citation2011, Citation2014). In Scenedesmaceae, the tufA and ITS genes appear to be the most practical combination for sufficient phylogenetic signal and ease of use.
Author contributions
G. Liu and Z. Hu: original concept; Q. Wang: drafting and editing manuscript; Q. Wang and X. Liu: analysis of molecular data and microscopic observation; Q. Wang, S. Li and Q. Xiong: culture of the strain and electron microscopy.
TEJP-2019-0144-File012.pdf
Download PDF (274.1 KB)TEJP-2019-0144-File011.pdf
Download PDF (329.2 KB)TEJP-2019-0144-File010.pdf
Download PDF (195 KB)TEJP-2019-0144-File009.pdf
Download PDF (221.6 KB)Acknowledgements
This work was supported by the National Natural Science Foundation of China (grants no. 31870189 and no. 31900187), Shanxi Province Science Foundation for Youths (201901D211421), Shanxi Key Laboratory for Research and Development of Regional Plants (SKL-RDRP), the Fund for Shanxi ‘1331 Project’ Key Innovative Research Team and Key Laboratory of Ecological Restoration of Water Pollution (Taiyuan Normal University). We thank the editor and reviewers for their helpful comments.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2020.1737968
Supplementary table 1. Strains whose sequences were downloaded from the GenBank.
Supplementary table 2. The h-CBCs and CBCs matrix of species in Hariotina (h-CBCs/CBCs).
Supplementary fig. 1. Morphology of Hariotina polychorda. Scale bar = 10 μm.
Supplementary fig. 2. Maximum-likelihood phylogenetic tree inferred from 18S sequences. Only bootstrap values above 50 and Bayesian posterior probabilities above 0.75 are shown. Boxes of the same colour represent the same species in Hariotina. Serial number represents intron insertion type. The strains provided in this study are indicated in bold.
References
- Bhattacharya, D., Friedl, T. & Damberger, S. (1996). Nuclear-encoded rDNA group I introns: origin and phylogenetic relationships of insertion site lineages in the green algae. Molecular Biology and Evolution, 13: 978−989.
- Caisová, L., Marin, B. & Melkonian, M. (2011). A close-up view on ITS2 evolution and speciation – a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evolutionary Biology, 11: 262.
- Dangeard, P.A. (1889). Mémoire sur les algues. Le Botaniste, 1: 127−174.
- Darty, K., Denise, A. & Ponty, Y. (2009). VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics, 25: 1974–1975.
- Famà, P., Wysor, B., Kooistra, W.H. & Zuccarello, G.C. (2002). Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene1. Journal of Phycology, 38: 1040−1050.
- Friedl, T., Besendahl, A., Pfeiffer, P. & Bhattacharya, D. (2000). The distribution of group I introns in lichen algae suggests that lichenization facilitates intron lateral transfer. Molecular Phylogenetics and Evolution, 14: 342−352.
- Fučíková, K., Rada, J.C., Lukešová, A. & Lewis, L.A. (2011). Cryptic diversity within the genus Pseudomuriella Hanagata (Chlorophyta, Chlorophyceae, Sphaeropleales) assessed using four barcode markers. Nova Hedwigia, 93: 29−46.
- Fučíková, K., Lewis, P.O. & Lewis, L.A. (2014). Putting incertae sedis taxa in their place: a proposal for ten new families and three new genera in Sphaeropleales (Chlorophyceae, Chlorophyta). Journal of Phycology, 50: 14−25.
- Guiry, M.D. & Guiry, G.M. (2020). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 11 February 2020.
- Hegewald, E., Coesel, P.F.M. & Hegewald, P. (2002). A phytoplankton collection from Bali, with the description of a new Desmodesmus species (Chlorophyta, Scenedesmaceae). Algological Studies, 105: 51−78.
- Hegewald, E., Wolf, M., Keller, A., Friedl, T. & Krienitz, L. (2010). ITS2 sequence-structure phylogeny in the Scenedesmaceae with special reference to Coelastrum (Chlorophyta, Chlorophyceae), including the new genera Comasiella and Pectinodesmus. Phycologia, 49: 325−335.
- Hoshaw, R.W. & Rosowski, J.R. (1973). Methods for microscopic algae. In Handbook of Phycological Methods. Culture Methods and Growth Measurements (Stein, J., editor), 53–67. Cambridge University Press, New York.
- Katoh, K. & Standley, D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30: 772−780.
- Kaufnerová, V. & Eliáš, M. (2013). The demise of the genus Scotiellopsis Vinatzer (Chlorophyta). Nova Hedwigia, 97: 415−428.
- Koršikov, O.A. (1953). Pidklas Protokokovi (Protococcineae): Vakuolʹni (Vacuolales) ta Protokokovi (Protococcales). Vydavnyctvo Akad. Nauk Ukraïnsʹkoï RSR.
- Krienitz, L., Hegewald, E., Hepperle, D. & Wolf, M. (2003). The systematics of coccoid green algae: 18S rRNA gene sequence data versus morphology. Biologia, Bratislava, 58: 437−446.
- Liu, X., Zhu, H., Song, H., Wang, Q., Xiong, X., Wu, C., Liu, G. & Hu, Z. (2018). Euchlorocystis gen. nov. and Densicystis gen. nov., two new genera of Oocystaceae algae from high-altitude semi-saline habitat (Trebouxiophyceae, Chlorophyta). Journal of Eukaryotic Microbiology, 65: 200−210.
- Lu, G. & Moriyama, E.N. (2004). Vector NTI, a balanced all-in-one sequence analysis suite. Briefings in Bioinformatics, 5: 378−388.
- Medlin, L., Elwood, H.J., Stickel, S. & Sogin, M.L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71: 491−499.
- Nylander, J.A. (2004). MrModeltest v2 (Program distributed by the author, Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden).
- Printz, H. (1927). Chlorophyceae (nebst Conjugatae, Heterocontae und Charophyta). In Die Naturlichen Pflanzenfamilien nebst ihren Gattungen und wichtiger Arten insbesondere der Nutzpflanzen, 3. Band (A. Engler & K. Prantl, editors). Wilhelm Engelmann, Leipzig.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A. & Huelsenbeck, J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539−542.
- Schultz, J., Müller, T., Achtziger, M., Seibel, P.N., Dandekar, T. & Wolf, M. (2006). The internal transcribed spacer 2 database – a web server for (not only) low level phylogenetic analyses. Nucleic Acids Research, 34: W704−W707.
- Sciuto, K., Lewis, L.A., Verleyen, E., Moro, I. & La, R.N. (2015). Chodatodesmus australis sp. nov. (Scenedesmaceae, Chlorophyta) from Antarctica, with the emended description of the genus Chodatodesmus, and circumscription of Flechtneria rotunda gen. et sp. nov. Journal of Phycology, 51: 1172−1188.
- Senn G. (1899). Über einige coloniebildende einzellige Algen. Botanische Zeitung, 57: 39−105.
- Seibel, P. N., Müller, T., Dandekar, T., Schultz, J. & Wolf, M. (2006). 4SALE – a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics, 7: 498.
- Seibel, P.N., Müller, T., Dandekar, T. & Wolf, M. (2008). Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Research Notes, 1: 91.
- Smith, G.M. (1920). Phytoplankton of the inland lakes of Wisconsin. Wisconsin Geological and Natural History Survey, Bulletin, 57: 1−234.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312−1313.
- Stanier, R.Y., Kunisawa, R., Mandel, M. & Cohen-Bazire, G. (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriological Reviews, 35: 171−205.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30: 2725−2729.
- Tavera, R. & Calderon, E.V.A. (2013). Use of CTAB as a cost-effective solution to an old problem: the interference of the mucilage of desmids for scanning electron microscopy. Phycologia, 52: 422−425.
- Thiers, B. (2019). Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/.
- Wang, Q., Song, H., Liu, X., Liu, B., Hu, Z. & Liu, G. (2019). Morphology and molecular phylogeny of coccoid green algae Coelastrella sensu lato (Scenedesmaceae, Sphaeropeales), including the description of three new species and two new varieties. Journal of Phycology, 55: 1290−1305.
- Wille, N. (1909). Conjugatae und Chlorophyceae. In Die natürlichen Pflanzenfamilien (A. Engler & K. Prantl, editors), Nachträge zum 1. Teil. Engelmann, Leipzig. 136 pp.