ABSTRACT
Oocystis-like algae are diverse and found throughout the world. In this study, two new strains of algae with colonies resembling Oocystis were identified and successfully cultured in the laboratory. A polyphasic approach, including light microscopy, transmission electron microscopy and phylogenetic analyses of 18S rDNA and rbcL cpDNA, was used to characterize the strains. Observation of the two strains showed a regular colonial morphology (cells/daughter colony are parallel to each other and positioned near the edge of the mother cell wall) and a characteristic fibrillar network structure on the mother cell wall is obvious in light microscopy, which distinguished them from other Oocystis-like members described previously. Molecular analyses revealed that the two strains form an independent lineage within the subfamily Makinoelloideae (Oocystaceae, Trebouxiophyceae), separated from other genera. Therefore, we described this alga as Reticulocystis yunnanensis gen. et sp. nov. The genera of Makinoelloideae with similar colonial morphotypes were revealed as not monophyletic and the relationships between them remained undetermined. Additional taxon sampling and data from additional multiple genes are necessary to assess the taxonomy of the Makinoelloideae.
Introduction
The family Oocystaceae is a well-supported clade in the Trebouxiophyceae (Chlorophyta) that is well known in aquatic ecosystems (Krienitz & Bock, Citation2011). The characteristic cell wall is composed of multiple cellulose layers with perpendicular fibril orientations. The mother cell walls often expand and retain the daughter cells forming specific Oocystis-like colonies (Komárek & Fott, Citation1983; Hepperle et al., Citation2000). The Oocystaceae has traditionally been considered to comprise four subfamilies, Lagerheimioideae, Oocystoideae, Eremosphaeroideae and Glaucocystidoideae (Komárek & Fott, Citation1983; Hepperle et al., Citation2000). Recent studies proposed three subfamilies (Eremosphaeroideae, Oocystoideae and Makinoelloideae) based mainly on molecular phylogeny combined with morphological methods (Štenclová et al., Citation2017).
The recently erected subfamily Makinoelloideae with the type genus Makinoella Okada was characterized by oval, elliptical or elongated cells arranged in pairs or tetrads to form multicellular coenobia or para-filamental fibres (Štenclová et al., Citation2017). So far, seven species from six genera, Makinoella tosaensis Okada, Willea vilhelmii (Fott) Komárek, Willea rectangularis (A.Braun) D.M.John, M.J.Wynne & P.Tsarenko, Elongatocystis ecballocystiformis Krienitz & C.Bock, Ecballocystis hubeiensis Hu & Liu, Ecballocystopsis dichotomus Hu & Bi and Oocystis nephrocytioides Fott & Čade, have been placed in this subfamily (Hepperle et al., Citation2000; Krienitz & Bock, Citation2011; Xia et al., Citation2013; Li et al., Citation2014; Štenclová et al., Citation2017). These taxa fall into three morphological types, i.e. crucigenioid four-celled, para-filamental fibres and Oocystis-like colonies.
The majority of the Makinoelloideae, including M. tosaensis, W. vilhelmii and W. rectangularis, have a typical Crucigenia-like four-celled coenobial morphology, which means that four cells of one generation live together for all their life and are arranged in a cross shape in more or less one plane (Komárek, Citation1974; Bock et al., Citation2013). In coenobia, each cell touches at least two other cells and the gap in the middle may or may not be present, in the latter case all four cells are in touch. Compound coenobia occur sometimes which means that more than one generation of cells in tetrads continue living together. M. tosaensis, as the only representative species of the genus Makinoella, was originally described from Japan (Okada, Citation1949) and later reported from Korea (Hegewald et al., Citation1999), Slovakia (Hindák & Hindákova, Citation2010) and China (Zhang et al., Citation2016). The species was originally placed in the family Oocystaceae, a placement supported by Bourrelly (Citation1966). Subsequently, based on colony formation, Komárek & Fott (Citation1983) transferred the family to the subfamily Crucigenioideae (Scenedesmaceae). It was since transferred back to the Oocystaceae by Hepperle et al. (Citation2000) based on molecular analyses. The genus Willea Schmidle, another quadrately four-celled coenobial member of the Makinoelloideae, was described on the basis of Crucigenia irregularis Wille as W. irregularis. Willea shares a similar morphology with Crucigeniella Lemmermann, which was erected in the same year (1900). Later, Lemmermann transferred the type of the genus Willea, Willea irregularis (Wille) Schmidle, to the genus Cohniella Schröeder (Lemmermann, Citation1904). However, this was not accepted by Komárek (Citation1974) who recognized a second species, Willea vilhelmii (Fott) Komárek, which was originally placed in the genus Dispora Printz by Fott (Citation1933). Komárek & Fott then included both Willea and Crucigeniella into the subfamily Crucigenoideae (Scenedesmaceae) (Komárek & Fott, Citation1983; Štenclová & Fučíková, Citation2019). Subsequently, John & Tsarenko (Citation2002) considered the occasional 90° alignment of daughter cells to the long axis of the mother coenobium as insufficient to separate these two genera, and merged Willea with Crucigeniella. However, Crucigeniella was illegitimate because it is a later homonym of a name validated earlier by Gaillon (Citation1833). Therefore, all Crucigeniella species were transferred to Willea (John et al., Citation2014). Up to now, two of the 10 accepted species listed in AlgaeBase (Guiry & Guiry, Citation2019), W. vilhelmii and W. rectangularis, have been sequenced and placed in the Makinoelloideae (Krienitz et al., Citation2003; Bock et al., Citation2013; Štenclová et al., Citation2017).
Differing considerably from the quadrately four-celled coenobia are two recent additions to the family Oocystaceae, the genera Ecballocystis Bohlin and Ecballocystopsis Iyengar, which exhibit para-filamental morphology. Ecballocystis was established by Bohlin (Citation1897) with the type species Ecballocystis pulvinata Bohlin. The genus was originally ascribed to the Chlorodendraceae (Tetrasporales), with a series of subsequent taxonomic rearrangements and, until its molecular phylogeny was determined (Xia et al., Citation2013), Komárek & Fott (Citation1983) included it in the Botryococcaceae (order Chlorococcales). Until now, 10 accepted species have been described (Guiry & Guiry, Citation2019). Finally, one strain of the species Ecballocystis hubeiensis was sequenced and positioned in the Oocystaceae (Xia et al., Citation2013), in agreement with Iyengar’s (Citation1932) conclusions. The genus Ecballocystopsis was established by Iyengar (Citation1933) with type species E. indica Iyengar. It was originally assigned to the Chlorodendrales and then moved to the Botryococcaceae with Ecballocystis by Komárek & Fott (Citation1983). Only one species, E. dichotomus, has been sequenced so far (Guiry & Guiry, Citation2019; Xia et al., Citation2013). In the Oocystaceae, Ecballocystis and Ecballocystopsis are two rare genera with para-filamentous morphology and differ from other para-filaments, Planctonema-like members, in the bigger cell size (20–45 μm long and 7–14 μm wide), more chloroplasts (16–24) and the character of filament formation (several generations of daughter cells enclosed in an expanded mother cell wall) (Liu et al., Citation2017). Ecballocystopsis differs from Ecballocystis in being simple para-filamentous instead of dendroid para-filamentous (Xia et al., Citation2013). Unlike most species of Oocystaceae living in aquatic environments, members of these two genera grow on the surface of wet substrata (Xia et al., Citation2013).
The remaining two species of Makinoelloideae: Elongatocystis ecballocystiformis (previously Oocystis ecballocystiformis) and O. nephrocytioides, share the Oocystis-like morphology of the colony and therefore were previously both positioned in the subfamily Oocystoideae within the Oocystaceae (Komárek & Fott, Citation1983). In contrast to the crucigenoid four-celled coenobial and para-filament members in Makinoelloideae mentioned above, cells of these two species are arranged loosely in the expanded mother cell wall.
In this study, two new strains with Oocystis-like colonial morphology were isolated in China and Indonesia. Morphological and phylogenetic analyses identified these strains as belonging to a new genus, Reticulocystis gen. nov., in the Makinoelloideae (Oocystaceae, Trebouxiophyceae, Chlorophyta).
Materials and methods
Samples of Reticulocystis yunnanensis were collected from artificial ponds of Xishuangbanna tropical botanical garden of the Chinese Academy of Sciences (strain LXD111, 21°55′N, 101°15′E, alt. 546 m, Yunnan Province, China, in August 2016) and another artificial pond in Monkey Forest (strain LXD107, 8°31′S, 115°15′E, alt. 190 m, Bali, Indonesia, in November 2016) by the authors. Both strains were isolated using the serial dilution pipetting technique (Hoshaw & Rosowski, Citation1973) until single colonies were obtained. Subsequently, individual colonies were cultivated in liquid BG11 medium (Stanier et al., Citation1971) with a constant light source of 30–50 μmol photons m–2 s–1 and a temperature of 25°C.
Morphological investigations were carried out using an Olympus BX53 light microscope (Olympus Corp., Tokyo, Japan) equipped with differential interference contrast (DIC). Microphotographs were taken with an Olympus BX53 camera. For transmission electron microscopy (TEM), cells were fixed in 3% glutaraldehyde in 0.1 M cacodylate buffer and fixed in 1% aqueous OsO4 in 0.1 M cacodylate buffer, dehydrated in acetone and embedded in Spurr’s resin. Ultrathin sections were stained with uranyl acetate and lead citrate (Reynolds, Citation1963).
Algal cells were broken with mini beads in a bead beater (3110BX, Biospec Products, Bartlesville, USA). Genomic DNA was extracted using a Universal DNA Isolation Kit (AxyPrep, Suzhou, China). Primers and PCR conditions for the 18S rDNA and rbcL cpDNA genes were previously described in Xia et al. (Citation2013).
The newly obtained sequences of the two strains were aligned with sequences of representative taxa of Trebouxiophyceae with focus on Oocystaceae (obtained from GenBank NCBI) on the basis of previous publications (Hepperle et al., Citation2000; Krienitz & Bock, Citation2011; Xia et al., Citation2013; Štenclová et al., Citation2017) controlled by BLAST and completed by all informative sequences from GenBank (National Center for Biotechnology Information [NCBI] http://www.ncbi.nlm.nih.gov/). The sequences were initially aligned using ClustalX version 2.0 (Larkin et al., Citation2007) and, after visual inspection, edited manually in Mega version 5 (Tamura et al., Citation2011). The 18S rDNA and rbcL cpDNA sequence positions that could not be aligned with confidence were removed prior to the analysis. Gaps were treated as missing data. Phylogeny was estimated using Maximum likelihood (ML) in RAxML version 8.0 (Stamatakis, Citation2014) and Bayesian inference (BI) in MrBayes 3.1.2 (Huelsenbeck & Ronquist, Citation2001). GTRGAMMA was selected as the best-fitting model for ML analysis inferred via RAxML version 8.0 (Stamatakis, Citation2014). Two different substitution models for each dataset were selected by MrModeltest 2.3 (Nylander, Citation2004) for the BI analyses: GTR+I+G for the 18S rDNA dataset and TIM+I+G for the rbcL cpDNA dataset. All Monte Carlo Markov Chain (MCMC) analyses were performed with four Markov chains (three heated chains, one cold) for 3×106 generations, where one tree was sampled every 1000 generations. Each analysis reached stationarity (an average standard deviation of split frequencies between runs < 0.01) well before the end of the run. The first 25% of the trees was discarded as burn-in before constructing the majority rule consensus trees in MrBayes.
Results
Morphological observations
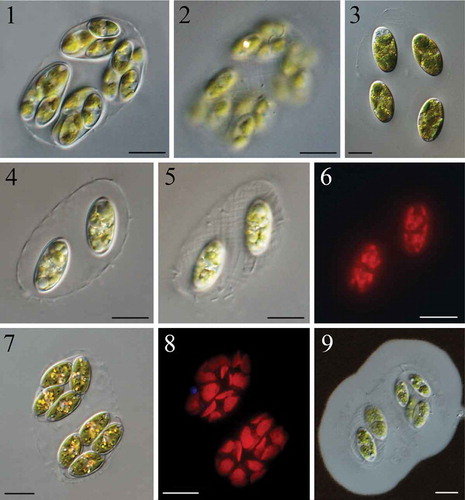
Colonies of 2–4 cells or composite colonies with 2–3 generations of cells were microscopic, enclosed in the mother cell walls and seldom found solitary in the field (, ). However, when cultured, solitary cells were more frequent than colonies (). Cells in the colony or daughter colonies in composite colonies were regularly parallel and positioned near the edge of the expanded mother cell wall (, , , ). The cells/daughter colonies were often aligned with their mother cell wall (when a 4-cell colony, , , ) or at a 45° angle to it (when a 2-cell colony, , ). The mucilage envelope around the colony was ~10 μm thick (). Mother cell walls usually had an elongated and irregular elliptical shape, with a clearly visible fibrillar network covering the entire surface in both field-collected and cultured material (, , , ). The fibrils were oriented spirally at an angle of 45° to the length of the mother cell wall. Cells were oval to elongated, elliptical with round ends or rare small tip thickenings, 12.60–25.08 μm long and 6.06–15.09 μm wide (, ). Young cells had two or four parietal chloroplasts, fragmenting into pieces with oil droplets inside old cells (, , ). Each chloroplast contained a central pyrenoid, often indistinct and not easy to observe in light microscopy (, , , ). Numerous oil droplets and other assimilated particles were a common occurrence inside matured cells (). Propagation was by oblique division forming 2–4 autospores (, ). Young daughter cells were tightly attached to each other and, separating slowly, still adhered to the mother cell wall during their maturation (, , ). The mother cell wall slowly gelatinized and eventually released the daughter cells ().
Figs 1–9. Light microscopy of Reticulocystis yunnanensis: Fig. 1. Characteristic composite colony morphology in field-collected sample. Fig. 2. Details of mother cell wall of composite colony show the obvious fibrillar network structure in field-collected sample. Fig. 3. Characteristic simple colony when cultured. Fig. 4. Characteristic simple colony in natural sample. Fig. 5. Details of mother cell wall of simple colony showing the fibrillar network structure in field-collected sample. Fig. 6. Autofluorescence shows the shape of chloroplasts in old cells. Fig. 7. Mature cells with numerous assimilate particles. Fig. 8. Autofluorescence shows the shape of chloroplasts in mature cells. Fig. 9. Negative staining with Indian ink showing mucilaginous envelopes. Scale bar: Figs 1–9, 10 μm
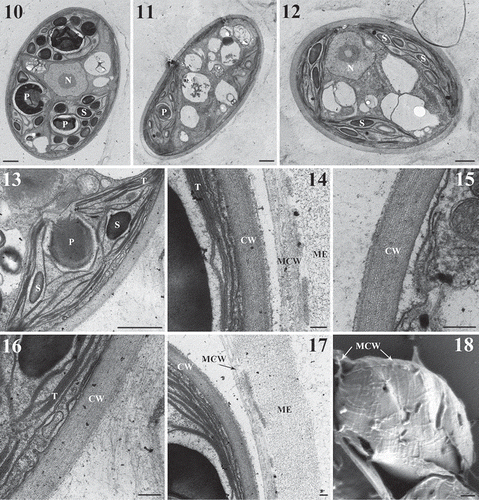
When observed with TEM, a multilayered cell wall and a thick mucilage envelope were obvious (–). One or two globular pyrenoids with a homogeneous matrix were situated in each chloroplast and surrounded by a starch sheath (–). Thylakoids extended over the length of the chloroplast and occurred in stacks of two to five (, ). Some of the tubular thylakoids penetrated the pyrenoid matrix (). Starch grains were numerous inside the chloroplast (, ). The fibrillar network structure of the cell wall was clearly visible on the collapsed mother cell wall in scanning electron microscopy ().
Figs 10–18. Transmission electron microscopy (Figs 10–17) and scanning electron microscopy (Fig. 18) of Reticulocystis yunnanensis. Fig. 10. Mature cell showing central pyrenoids in each chloroplast. Fig. 11. Mature cell showing oil droplets. Fig. 12. Mature cell with numerous starch grains inside the chloroplast. Fig. 13. Details of pyrenoids penetrated by tubular thylakoids. Fig. 14. Details of mother cell wall. Fig. 15. Details of multi-layered cell wall. Fig. 16. Details of cell wall and chloroplast thylakoids. Fig. 17. Details of mucilage envelope. Fig. 18. Mother cell wall collapsed on the daughter cell shows the details of fibrillar network structure. (CW = cell wall, MCW = mother cell wall, ME = mucilage envelope, P = pyrenoid, S = starch grains, T = thylakoids, and N = nucleus). Scale bar: Figs 10–13, 18, 1 μm; Figs 15–16, 0.2 μm; Figs 14, 17, 0.1 μm
Phylogenetic analyses
18S rDNA and rbcL cpDNA sequences were obtained for both strains. Sequencing of the 18S rDNA PCR product of R. yunnanensis produced a 2521 bp sequence containing two introns (LXD107) and a 2628 bp sequence with three introns (LXD111). Introns were not found in rbcL cpDNA sequences. Sequences obtained herein were submitted to GenBank under accession numbers MN334212–MN334217.
The final alignment of the 18S rDNA exon regions included the main taxa in the Trebouxiophyceae based on 1474 bp. Ankistrodesmus fusiformis (Chlorophyceae) was chosen as the outgroup. The aligned rbcL cpDNA dataset with 1037 bp included mainly Oocystaceae and members of the Chlorellaceae were chosen as the outgroup.
The ML and Bayesian analyses yielded similar topologies and the ML is presented. The 18S rDNA phylogenetic trees and the rbcL cpDNA trees both recovered the new genus Reticulocystis in the Makinoelloideae (Oocystaceae). The genus Reticulocystis formed an early-diverging subclade within the main clade of the Makinoelloideae (containing all taxa other than Ecballocystopsis) with maximum statistical support in 18S rDNA trees (). However, in the rbcL cpDNA analyses, Reticulocystis was recovered in a different position within the Makinoelloideae, forming a lineage sister to a clade including members of Willea and Makinoella, but with low statistical support (0.71/52 for BI/ML) (). The genus Elongatocystis was absent in the rbcL cpDNA analyses and therefore its relationship to Reticulocystis could not be clarified.
Discussion
In the small artificial ponds of the Xishuangbanna tropical botanical garden of the Chinese Academy of Sciences, members of Oocystaceae were found frequently throughout the year and were very diverse especially in spring and summer, for example species of Makinoella (Zhang et al., Citation2016), Eremosphaera De Bary, Oocystis Nägeli ex A. Braun, Oocystidium Korshikov, Echinocoleum C.-C. Jao & K.T.Lee, Lagerheimia Chodat and Quadricoccopsis X.Liu, H.Zhu, H.Song, B.Liu, Q.Wang, G.Liu & Z.Hu (Liu et al., Citation2018). Here, a new member of this group was also found in these ponds and in another similar habitat in Bali. An integrative taxonomic study including molecular analyses revealed it as a new genus, Reticulocystis, in the Makinoelloideae (Oocystaceae).
The morphology of the genus Reticulocystis can be characterized as Oocystis-like colonies, one of the morphology types in Makinoelloideae, which includes also Oocystis nephrocytioides and Elongatocystis ecballocystiformis, and is distinguished from the other types by obviously different colony morphology and cell arrangement. Unlike O. nephrocytioides, R. yunnanensis can be distinguished by its non-asymmetrical cell shape, often more than two-celled colonies and mother cell wall not completely dissolving during formation of autospores (Fott & Čado, Citation1966). Comparatively, the new species is more similar to E. ecballocystiformis, which was described by Iyengar (Citation1932) as O. ecballocystiformis. Then, because of the distant phylogenetic position from other Oocystis species, Krienitz & Bock (Citation2011) excluded it from Oocystis and described the new genus Elongatocystis based on it. Elongatocystis can differ from Reticulocystis by the transverse division during propagation, which is an important feature mentioned by Iyengar (Citation1932) to distinguished Elongatocystis from loose cells of Ecballocystis fritschii M.O.P.Iyengar. In addition, Elongatocystis was described as solitary and occasionally forming colonies with 2–4–8 daughter cells (Iyengar, Citation1932), which are different from the usually 2–4-celled colonies or bigger composite colonies with 2–3 generations found in Reticulocystis. In culture, a mucilage envelope is lacking in Elongatocystis (Krienitz & Bock, Citation2011) but thick in Reticulocystis. Furthermore, cells in colonies of Elongatocystis are loosely enclosed in the mother cell wall (Iyengar, Citation1932). Conversely, in Reticulocystis, cells are regularly arranged parallel or at a 45° angle to the length of the mother cell wall and the daughter cells/colonies often are close to the edge of their expanded mother cell wall. This typical cell arrangement in the colonies is distinctive in all Oocystis-like species in the Oocystaceae. Although the colony morphology of Oocystis is diverse, according to Komárek & Fott (Citation1983), the parallel-arranged cells/daughter colonies were only mentioned in O. ecballocystiformis var. americana Komárek, a variant of E. ecballocystiformis described by Komárek (Citation1983). This variant, however, differed from Reticulocystis by the centrally positioned daughter cells in the colonies. The character of the parallel cells/daughter colonies arrangement is more common in the genera Willea, Ecballocystis and Ecballocystopsis, which may imply their close evolutionary relationships in the Makinoelloideae.
In addition, a fibrillar network structure on the mother cell wall is a striking feature of R. yunnanensis. The crosswise microfibrillar structure on the cell wall is common in the Oocystaceae and has been observed by TEM in Oocystis (Robinson & Preston, Citation1972), Lagerheimia (Hegewald et al., Citation1978), Siderocelis (Naumann) Fott (Crawford, Citation1978), Franceia Lemmermann (Hegewald et al., Citation1980), Siderocystopsis E.M.F. Swale (Hegewald & Schnepf, Citation1984) and Makinoella (Hegewald et al., Citation1999). Compared with previous observations, the fibrillar network structure of Reticulocystis seems looser and thicker, observable even in light microscopy without any treatment. Hindák & Hindakova (Citation2010) mentioned clearly visible radiating fibrils or radially arranged endogloeic rod-like bacteria in Makinoella tosaensis. Rod-like bacteria were also often visibly randomly distributed on the mother cell wall of R. yunnanensis (–). Different to the radiating fibrils or radially arranged rod-like bacteria described for M. tosaensis, in R. yunnanensis the fibrillar network structure was tiled and more regular. On the other hand, in multi-generational composite colonies, the fibrillar network structure can also be observed on the mother cell wall of the inner daughter colonies, which are enclosed within the old mother cell wall and not easily invaded by bacteria (). Whether the type of fibrillar network structure is specific in different taxa of Oocystaceae or affected by environmental conditions, or at which level it is taxonomically significant, we cannot conclude here and more research is needed. We have, however, been able to distinguish two morphologically and phylogenetically close genera, e.g. Reticulocystis and Elongatocystis.
The phylogenetic position of R. yunnanensis was inconsistent in the 18S rDNA and rbcL cpDNA analyses. It is not uncommon in phylogenetic analysis of green algae that different single gene-phylogenies yield different topologies (Krienitz et al., Citation2004; Fučíková et al., Citation2014; Štenclová et al., Citation2017), particularly if the compared markers have different substitution rates, as is the case for 18S rDNA and rbcL cpDNA. For R. yunnanensis, the phylogenetic position was fully resolved by 18S rDNA, which placed this species as sister to the main clade of Makinoelloideae, containing all other taxa except Ecballocystopsis (with the morphologically similar Elongatocystis ecballocystiformis recovered as the subsequent diverging taxon within the main clade of Makielloideae). The rbcL cpDNA dataset lacks E. ecballocystiformis, a gap that certainly contributes to the unresolved position of R. yunnanensis.
Compared with the common large genera in the Oocystaceae, especially the broadly defined Oocystis which may need a thorough revision (Krienitz & Bock, Citation2011), Makinoelloideae is composed of a limited number of small genera with distinct morphological and molecular differences. Both 18S rDNA and rbcL cpDNA resolved the seven genera (six original genera and the new genus) of Makinoelloideae into six clades, i.e. Ecballocystopsis clade, Reticulocystis clade, Elongatocystis clade, Willea clade, Makinoella clade and Ecballocystis/Oocystis nephrocytioides clade. However, the three morphotypes (mentioned in the Introduction) do not cluster in monophyletic clades. The para-filamentous Ecballocystopsis is the earliest-diverging lineage in the subfamily and is well separated from the main clade, which includes another para-filamentous member (Ecballocystis) that formed a well-supported subclade with O. nephrocytioides in both phylogenies. O. nephrocytioides, which requires a similar taxonomic revision as was performed for Elongatocystis ecballocystiformis by Krienitz & Bock (Citation2011) for the same reason of its distant phylogenetic position from other Oocystis species, is also not clustered with the other two genera with Oocystis-like colony morphology (Reticulocystis and Elongatocystis). The phenomenon that strains with similar morphology do not represent a natural phylogenetic group is common in Oocystaceae. The genus Oocystis was polyphyletic and scattered through almost the whole clade of Oocystaceae (Krienitz & Bock, Citation2011). Oocystidium planoconvexa (Hindák) Štenclová & Pažoutová and Echinocoleum elegans C.-C. Jao & K.T.Lee, with similar ray-like mucilaginous envelopes, are also not closely related (Pažoutová et al., Citation2010; Štenclová et al., Citation2017). The easily confused, slender filaments of Planctonema-like members were distributed into three genera, and Planctonemopsis Liu, Zhu, Liu, Hu & Liu was far from the other two (Liu et al., Citation2017). A similar case was shown for Quadricoccus Fott and Quadricoccopsis, which are both characterized by oval to ellipsoid cells adherent to the bowl-shaped or stretched, empty mother cell walls (Liu et al., Citation2018). Chlorellaceae and the crucigenioid algae shared a similar situation (Wolf et al., Citation2002; Krienitz et al., Citation2004; Luo et al., Citation2010; Pröschold et al., Citation2010; Bock et al., Citation2011, Citation2013). These results point to the need to explore the phenotypical evolutionary relationships of these clades in the future. Apart from the early-diverging Ecballocystopsis, the phylogenetic relationships among the other Makinoelloideae have not yet been resolved fully, a situation common to the Oocystaceae (Štenclová et al., Citation2017). Many small clades in the Oocystoideae are weakly supported and the reason for such low support is not easy to understand. Low phylogenetic signal was detected in the 18S rDNA dataset and sequences of rbcL cpDNA are still missing for many taxa. Advanced taxon sampling and data from additional molecular markers are needed in the future.
Taxonomic assessment
Reticulocystis X.D. Liu, G.X. Liu & Z.Y. Hu, gen. nov.
Diagnosis: 2–4 cells in colonies and sometimes composite colonies formed by 2–3 generations, seldom solitary cells. Mother and grandmother cell walls with a characteristic fibrillar network structure covering the entire surface. In the colony, cells/daughter colonies with regularly parallel arrangement or at a 45° angle to the mother cell wall. Mucilage envelope around colony thick. Cells oval to elliptical. 2–4 parietal chloroplasts, each with one pyrenoid. Cell division oblique, forming 2–4 autospores. The genus differs from other members of the Makinoelloideae in the regular cell/daughter colony arrangement (parallel to each other and positioned near the edge of the mother cell wall), the characteristic fibrillar network structure on the mother cell wall and the nucleotide sequences for 18S rDNA and rbcL cpDNA.
Etymology: The genus is named for the obvious fibrillar network fully covering the mother cell wall (Reticul-, from the Latin word reticulum = small net; and -ocystis, from cystis = cyst, derived from the ancient Greek κύστις = bladder, sac).
Type species: Reticulocystis yunnanensis.
Reticulocystis yunnanensis X.D. Liu, G.X. Liu & Z.Y. Hu, sp. nov.(Figs 1–18)
Diagnosis: Colonies formed by 2–4 cells or composite colonies formed by 2–3 generations. Cells oval to elongated elliptical with round ends, rare small tip thickenings, 12.60–25.08 μm long and 6.06–15.09 μm wide. Two or four parietal chloroplasts per cell, fragmenting into pieces with numerous assimilate particles and oil droplets when old. Propagation by 2–4 autospores.
Holotype: Formaldehyde-fixed material stored at the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China, as specimen No. LXD111.
Reference strain: A living culture deposited in the Freshwater Algae Culture Collection, Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China (FACHB) as strain FACHB-2376. Isolated from water samples collected in August 2016.
Type locality: Small artificial ponds of Xishuangbanna Tropical Botanical Garden of Chinese Academy of Sciences (21°55′N, 101°15′E, alt. 546 m, Mengla county, Yunnan Province, China).
Etymology: The species name indicates the collection locality of the type strain.
Author contributions
X. Liu: original concept, collection of the samples, experimental design and application, analyses of data, drafting and editing manuscript. Q. Wang: experimental design, electron microscopy and culture of the strains. H. Zhu: experimental design and analysis of molecular data. B. Liu: experimental design and culture of the samples. G. Liu: original concept, experimental design and editing of the manuscript. S. Xie: original concept and experimental design. Z. Hu: collection samples and experimental design. F. Rindi: language improvement.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (No. 31670202 and No. 31900187) and funded by the Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences (2018–002).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bock, C., Luo, W., Kusber, W.H., Hegewald, E., Pažoutová, M. & Krienitz, L. (2013). Classification of crucigenoid algae: phylogenetic position of the reinstated genus Lemmermannia, Tetrastrum spp., Crucigenia tetrapedia, and C. lauterbornii (Trebouxiophyceae, Chlorophyta). Journal of Phycology, 49: 329–339.
- Bock, C., Pažoutová, M. & Krienitz, L. (2011). Phylogenetic position of Coronastrum ellipsoideum and description of Parachlorella hussii sp. nov. Biologia, 66: 585–594.
- Bohlin, K. (1897). Die Algen der ersten Regnell’schen Expedition, 1: Protococcoideen. In Bihang till Svenska Vetenskapsakademie Handlingar, 23: 3–47. Stockholm.
- Bourrelly, P. (1966). Les algues d’eau douce, vol. 1. Les algues vertes. Boubée et Cie, Paris.
- Crawford, R.M. (1978). A new forma and a new species of the genera Scenedesmus and Siderocelis (Chlorophyceae) from the nanoplankton of a brackish-water coastal pool. Protoplasma, 96: 351–360.
- Fott, B. (1933). Die Schwebeflora des Ohrid-Sees. Bulletin de l’institut et du Jardin Botaniques de l’Universite de Beograd, 2: 153–175.
- Fott, B. & Čado, I. (1966). Oocystis nephrocytioides sp. nov. Phycologia, 6: 47–50.
- Fučíková, K., Lewis, P.O. & Lewis, L.A. (2014). Putting incertae sedis taxa in their place: a proposal for ten new families and three new genera in Sphaeropleales (Chlorophyceae, Chlorophyta). Journal of Phycology, 50: 14–25.
- Gaillon, B. (1833). Aperçu d’histoire naturelle et observations sur les limites qui séparent le règnevégétal du règne animal [suivi des] tableaux synoptiques et méthodiques des genres des Nématozoaires (Lu à la Société d’Agriculture, du Commerce et des Arts, de Boulogne-sur-mer, dans sa séance publique du 19 Septembre 1832). Imprimerie de Le Roy–Mabi, Boulogne.
- Guiry, M.D. & Guiry, G.M. (2019). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 31 July 2019.
- Hegewald, E. & Schnepf, E. (1984). On the structure and taxonomy of spiny Chlorellases (Micractiniaceae, Golenkiniaceae, Siderocystopsis). Nova Hedwigia, 34: 297–383.
- Hegewald, E., Schnepf, E. & Aldave, A. (1978). Investigations on the lakes of Peru and their phytoplankton. 4. The algae of Laguna Paca with special reference to Chodatella subsalsa and Scenedesmus ellipticus. Algological Studies/Archiv für Hydrobiologie, Supplement Volumes: 384–392.
- Hegewald, E., Schnepf, E. & Aldave, A. (1980). Investigations on the lakes of Peru and their phytoplankton. 5. The algae of Laguna Piuray and Laguna Huaypo, Cuzco, with special reference to Franceia, Oocystis and Scenedesmus. Algological Studies/Archiv für Hydrobiologie, Supplement Volumes: 387–420.
- Hegewald, E., Schnepf, E. & Jeon, S.L. (1999). Report on Makinoella tosaensis Okada (Chlorophyta, Oocystaceae), a species new to Korea. Algae, 14: 87–90.
- Hepperle, D., Hegewald, E. & Krienitz, L. (2000). Phylogenetic position of the Oocystaceae (Chlorophyta). Journal of Phycology, 36: 590–595.
- Hindák, F. & Hindakova, A. (2010). First report of Makinoella tosaensis OKADA (Chlorophyta, Chlorococcales, Oocystaceae) outside East Asia. Fottea, 10: 141–144.
- Hoshaw, R.W. & Rosowski, J.R. (1973). Methods for microscopic algae. In Handbook of Phycological Methods (Stein, J.R., editor). Cambridge University Press, New York.
- Huelsenbeck, J.P. & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17: 754–755.
- Iyengar, M.O.P. (1932). Two little-known genera of green algae (Tetrasporidium and Ecballocystis). Annals of Botany, 2: 191–192.
- Iyengar, M.O.P. (1933). Ecballocystopsis indica n. gen. et sp., a new member of Chlorodendrales. Annals of Botany, 47: 21–25.
- John, D.M. & Tsarenko, P.M. (2002). Order Chlorococcales. In The Freshwater Algal Flora of the British Isles (John, D.M., Whitton, B.A. & Brook, A.J., editors). Cambridge University Press, Cambridge.
- John, D.M., Wynne, M.J. & Tsarenko, P.M. (2014). Reinstatement of the genus Willea Schmidle 1900 for Crucigeniella Lemmermann 1900 nom. illeg. (Chlorellales, Trebouxiophyceae, Chlorophyta). Phytotaxa, 167: 212–214.
- Komárek, J. (1983). Contribution to the chlorococcal algae of Cuba. Nova Hedwigia, 37: 65–180.
- Komárek, J. (1974). The morphology and taxonomy of crucigenoid algae (Scenedesmaceae, Chlorococcales). Archiv für Protistenkunde, 116: 1–74.
- Komárek, J. & Fott, B. (1983). Chlorococcales. In Das Phytoplankton des Süßwassers (Huber-Pestalozzi, G., editor), Band 7. 1. Hälfte. 1044 pp. Schweizerbart’sche Verlagsbuchhandlung, Stuttgart.
- Krienitz, L. & Bock, C. (2011). Elongatocystis ecballocystiformis gen. et comb. nov., and some reflections on systematics of Oocystaceae (Trebouxiophyceae, Chlorophyta). Fottea, 11: 271–278.
- Krienitz, L., Hegewald, E., Hepperle, D. & Wolf, M. (2003). The systematics of coccoid green algae: 18S rRNA gene sequence data versus morphology. Biologia, 58: 437–446.
- Krienitz, L., Hegewald, E.H., Hepperle, D., Huss, V.A.R., Rohr, T. & Wolf, M. (2004). Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia, 43: 529–542.
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. & Higgins, D.G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23: 2947–2948.
- Lemmerman, E. (1904). XVII. Über de Entstehung neuer Planktonformen. Berichte der Deutschen Botanischen Gesellschaft, 22: 17–22.
- Li, F., Huang, X. & Li, C. (2014). A comparison and phylogenetic analysis of the pyrenoid ultrastructure of three Oocystis species (Oocystaceae, Trebouxiophyceae, Chlorophyta). Journal of Advances in Biology, 6: 861–867.
- Liu, X., Zhu, H., Liu, B., Liu, G. & Hu, Z. (2017). Classification of Planctonema‐like algae, including a new genus Planctonemopsis gen. nov., a new species Planctonema gelatinosum sp. nov. and a reinstated genus Psephonema (Trebouxiophyceae, Chlorophyta). Journal of Phycology, 53: 869–879.
- Liu, X., Zhu, H., Song, H., Liu, B., Wang, Q., Liu, G. & Hu, Z. (2018). Quadricoccopsis gen. nov., a new genus of Quadricoccus-like algae in Oocystaceae from China (Trebouxiophyceae, Chlorophyta). Fottea, 18: 189–199.
- Luo, W., Pröschold, T., Bock, C. & Krienitz, L. (2010). Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biology, 12: 545–553.
- Nylander, J.A.A. (2004). MrModeltest ver. 2.3. Program distributed by the author. Evolutionary Centre, Uppsala University, Sweden.
- Okada, Y. (1949). Makinoella tosaensis, a new genus of the Oocystaceae. Japanese Journal of Botany, 24: 166–168.
- Pažoutová, M., Škaloud, P. & Nemjová, K. (2010). Phylogenetic position of Ooplanctella planoconvexa, gen. et comb. nova and Echinocoleum elegans (Oocystaceae, Trebouxiophyceae, Chlorophyta). Fottea, 10: 75–82.
- Pröschold, T., Bock, C., Luo, W. & Krienitz, L. (2010). Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycological Research, 58: 1–8.
- Reynolds, E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. Journal of Cell Biology, 17: 208–212.
- Robinson, D.G. & Preston, R.D. (1972). Plasmalemma structure in relation to microfibril biosynthesis in Oocystis. Planta, 104: 234–246.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312–1313.
- Stanier, R.Y., Kunisawa, R., Mandel, M. & Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriological Reviews, 35: 171–205.
- Štenclová, L., Fučíková, K., Kaštovský, J. & Pažoutová, M. (2017). Molecular and morphological delimitation and generic classification of the family Oocystaceae (Trebouxiophyceae, Chlorophyta). Journal of Phycology, 53: 1263–1282.
- Štenclová, L & Fučíková, K. (2019). Dispora speciosa, a new addition to the genus Parallela and the first coccoid member of the family Microsporaceae. Phytotaxa, 419: 63–76.
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28: 2731–2739.
- Wolf, M., Krienitz, L. & Hepperle, D. (2002). Phylogenetic position of Actinastrum hatzschii Lagerheim 1882 (Chlorophyta, Trebouxiophyceae). Algological Studies, 104: 59–67.
- Xia, S., Zhu, H., Cheng, Y.Y., Liu, G.X. & Hu, Z.Y. (2013). Phylogenetic position of Ecballocystis and Ecballocystopsis (Chlorophyta). Fottea, 13: 65–75.
- Zhang, Q., Song, H., Zheng, L., Song, L. & Liu, G. (2016). Makinoella tosaensis Okada, a new recorded genus and species of freshwater Trebouxiophycean algae from China. Journal of Tropical and Subtropical Botany, 24: 406–412.