ABSTRACT
A new marine strain of picoplanktonic algae, PMPFPPE4, was isolated from a mixed net-phytoplankton sample taken from the upper euphotic layer of the southeastern Adriatic Sea. Evaluation of the new strain included morphological investigation (by light and electron microscopy), phylogenetic analysis (utilizing plastid 16S rRNA and nuclear 18S rRNA genes), and physiological characterization (screening of pigment/lipid composition and capturing photosynthesis measurements). The new strain was proven to belong to the genus Picochlorum and the lipid composition revealed an unexpected accumulation of triacylglycerols, indicating an evolutionary adaptation for growth under unfavourable conditions. In addition, lipid remodelling in the exponential to stationary growth phase was characterized by an increased share of membrane-forming digalactosyldiacylglycerols and phosphatidylcholines. Maximum photosynthetic activity measured was at 30°C, but the most rapid increase of photosynthetic activity was at lower temperatures (15–20°C). Moreover, the thermotolerant strain did not exhibit photoinhibition below 40°C and survived a one-month cultivation period in complete darkness. The strain’s survival in low light and dark conditions suggests a potential shift from autotrophy to mixotrophy under unfavourable growth conditions. Thus, the unique physiological attributes represented by a high growth rate, thermotolerance, phototolerance and high triacylglycerol synthesis may render the strain highly attractive for biofuel production and growth in large outdoor systems.
Introduction
Photosynthetic picoeukaryotes (PPEs), with cells less than 3 μm, represent a significant fraction of the picophytoplankton (Dı́ez et al., Citation2001). Although bacteria, cyanobacteria and archaea are dominant in abundance in the oceans, PPEs can greatly contribute to global carbon cycling due to their larger cell volume (Li, Citation1994; Richardson & Jackson, Citation2007).
Recently, PPE diversity research has been intensified by applying various molecular approaches and metabarcoding of environmental DNA (high-throughput sequencing of DNA markers), mostly variable regions of the nuclear 18S rRNA and plastid 16S rRNA gene, allowing better understanding of the diversity and importance of the pico-world (Fuller et al., Citation2006a, Citation2006b; Decelle et al., Citation2015; de Vargas et al., Citation2015). We owe today’s vast knowledge to cross-oceanic expeditions such as Malaspina, Tara Oceans, Biosope and Ocean Sampling Day (OSD) (Grob et al., Citation2007; Claustre et al., Citation2008; Bork et al., Citation2015; Duarte, Citation2015; de Vargas et al., Citation2015; Tragin & Vaulot, Citation2018). In addition to environmental metabarcoding approaches, cloning of environmental DNA, fingerprinting and tag-sequencing and single-cell genomics approaches are also being used to appraise protist diversity (Sieracki et al., Citation2019).
The most diverse eukaryotic organisms in the above-mentioned cross-oceanic expeditions always belong to the piconanoplankton, i.e. ultraplankton (cells with diameter ≤ 5 µm). Trebouxiophyceae, a class of green algae containing the highly diversified marine coccoid genus Picochlorum within the ‘core’ chlorophytes, comprise approximately one-third of the sequences in oceanic temperate areas and are more abundant in upwelling and in nutrient-rich coastal zones (Tragin et al., Citation2016). Recently, the genus Picochlorum was recognized as the most abundant trebouxiophyte in temperate areas of the USA and European Atlantic coast with the highest number of reads of major operational taxonomic units (OTUs) found in the OSD dataset (Tragin & Vaulot, Citation2018). In the Mediterranean, during the PROSOPE cruise, Dı́ez et al. (Citation2001) and Massana et al. (Citation2004) found a large diversity of PPEs, including putative photosynthetic representatives from a wide range of classes (e.g. Chrysophyceae, Cryptophyceae, Prasinophyceae and Prymnesiophyceae; Vaulot et al., Citation2008). Green algae in the southern Adriatic Sea comprised roughly 26% of photosynthetic reads, but the genus Picochlorum has not yet been confirmed among 18S rRNA reads (Tragin & Vaulot, Citation2018).
Algae culture is one of the most important methods for studying the diversity of these organisms and their role in the ecosystem, as well as for future applications. Physicochemical approaches can be useful in applied biology studies, such as biotechnology. At the simplest level, photosynthetic pigments (the key taxonomic diagnostic feature for microalgae) allow us to distinguish green, brown and red algae, but the photosynthetic pigment signature is often also indicative of the algal class (Guillou et al., Citation1999). Biotechnological approaches to algal studies mostly focus on lipids and fatty acids, since their target is often biofuel production (Hu et al., Citation2008; Liu et al., Citation2011). Algae that synthesize and accumulate large quantities of neutral lipids are good candidates for biofuels and biomaterials production (Hu et al., Citation2008). However, screening of algae for their cell lipid or fatty acid components can also tell us much about their lifestyle and environment (Galloway & Winder, Citation2015). Useful information about physiology may also emerge from photosynthesis measurements. The process of photosynthesis mainly depends on abiotic factors such as pH, carbon availability, light intensity and temperature (Kirk, Citation1994). The widely accepted P-I model describes the relationship between photosynthetic activity and light intensity (Webb et al., Citation1974; Platt et al., Citation1980) and provides parameters such as biomass-specific photosynthetic activity, photoinhibition, photoadaptation, light utilization or respiration. Since growth continues by the consumption of photosynthetic products, photosynthetic parameters can help us understand the behaviour of a species in the environment.
Here we describe a newly isolated pico green algal strain showing high competitiveness and survival in culture conditions. It was isolated together with pennate diatoms from the oligotrophic southern Adriatic Sea. During the fieldwork, a photosynthetic signal in the aphotic zone was noticed that was later recognized as representing photosynthetic picoeukaryotes (PPEs) in the dark at a depth of 280 m, while viable diatoms were observed up to 500 m (Bosak et al., Citation2016; Babić et al., Citation2017). When cultivating monoclonal PPEs and pennate diatoms, pico green algae (later recognized as different strains of Picochlorum, including PMFPPE4) often outgrew other cultures and survived harsh conditions (neglected and/or mixed culturing). We isolated it in monoclonal culture and characterized it by morphological analysis (using light and transmission electron microscopy), phylogenetic analysis (using plastid 16S rRNA and nuclear 18S rRNA genes), pigment and lipid screening, and photosynthesis measurements under different temperature and light regimes.
Materials and methods
Isolation and cultivation
The water sample was taken from 20 m depth to the surface during the 28 February–3 March 2015 BIOTA (Bio-tracing Adriatic Water Masses) cruise conducted in the south-eastern Adriatic Sea at station M300 (42.48N, 17.28E), using a 20 µm-mesh-size phytoplankton net. Environmental conditions and nutrient concentrations of the area are given in Babić et al. (Citation2017). Immediately upon arrival in the laboratory, the sample was inoculated in Guillard’s f/2 Marine Water Enrichment Solution (Sigma-Aldrich, UK). After a mixed culture had grown (containing mostly diatoms, dinoflagellates, cryptophytes, haptophytes and heterotrophic nanoflagellates), unknown PPE cells were filtered into a fresh medium through 3.0µm-pore-size Nucleopore polycarbonate membrane filters (Whatman, UK) with a syringe and a filter holder. Following filtration, isolation continued by the dilution method; repeated transfer of a sub-volume of a culture (1/10 of the medium volume) to a fresh medium (9/10 of the medium volume) in order to obtain one cell per tube at the end of the series (Knight-Jones, Citation1951; Throndsen, Citation1978). A xenic culture of strain PMFPPE4 was established and subsequently transplanted biweekly to fresh medium, thus keeping the cells in the exponential phase of growth. The strain was later cultivated in ASN III medium (Stanier et al., Citation1979), a synthetic medium used in photosynthetic experiments. The PMFPPE4 strain is deposited in the Roscoff Culture Collection under number RCC6905.
Morphology
The fresh culture samples were investigated under an Olympus BX51TF (Olympus Corporation, Japan) inverted microscope (Artray Co. Ltd, Japan) and a Zeiss Axioimager A2 light microscope (Carl Zeiss, Oberkochen, Germany) equipped with DIC and phase contrast, and an Axiocam 305 camera with which images were obtained. Before examination, PMFPPE4 cells were allowed to settle on glass slides for 20 min. The cells were examined using a 1000× magnification.
For transmission electron microscopy (TEM), cultured cells were fixed in 1% (w/v) glutaraldehyde in 50 mM cacodylate buffer (pH 7.2) for 30 min at 5°C and pelleted by centrifugation at 500 × g for 5 min. Cells were re-suspended with ice-cold 50 mM cacodylate buffer (pH 7.2) and embedded in 2% agarose. The agarose with the embedded cells was cut into small pieces and washed twice with ice-cold 50 mM cacodylate buffer (pH 7.2). The cells were then post-fixed with 1% (w/v) osmium tetroxide in the same buffer for 1 h at 4°C, followed by a 10 min wash in ice-cold water. After dehydration in a graded series of ethanol (25%, 50%, 75%, 80%, 90%, 100%), the material was placed in absolute ethanol overnight. On the next day the material was placed in a mixture of 1:1 absolute ethanol and 100% acetone for 30 min, then in 100% acetone for a further 30 min. The material was then placed in a mixture of Spurr’s medium and acetone: initially in ⅓ Spurr’s and ⅔ acetone for 30 min, followed by ½ Spurr’s and ½ acetone for 30 min and lastly in ⅔ Spurr’s and ⅓ acetone for 30 min. This was followed by placing the material in Spurr’s medium alone for 2 h at 45°C. Finally, the material was placed in a plastic mould and polymerized in Spurr’s medium at 65°C for 48 h. Ultrathin sections were made with an ultra-microtome Leica Ultracut R and stained with 4% aqueous uranyl acetate for 10 min, then with lead citrate, pH 12.0 for 10 min (Reynolds, Citation1963) and examined using a FEI Morgagni 268D transmission electron microscope (Eindhoven, the Netherlands) at 70 kV.
DNA extraction and PCR amplification
DNA extraction was performed with a DNeasy Plant Mini Kit (Qiagen), in accordance with manufacturer’s instructions, using 50 ml of PMFPPE4 culture in the exponential growth phase. Purity of the extracted DNA was assessed with the NanoDrop spectrophotometer (BioSpec-nano; Shimadzu, Tokyo, Japan). The plastid 16S rRNA and nuclear 18S rRNA genes were amplified by PCRs. For the 16S rRNA gene, we used algal plastid biased forward and reverse primers PLA491F (5’-GAGGAATAAGCATCGGCTAA-3’) (Fuller et al., Citation2006a) and OXY1313R (5’-CTTCAYGYAGGCGAGTTGCAGC-3’) (West et al., Citation2001), respectively. For the 18S rRNA gene, we used Euk63F (5’-CGCTTGTCTCAAAGATTA-3’) as forward and Euk1818R (5’-ACGGAAACCTTGTTACGA-3’) as reverse primers (Lepère et al., Citation2011).
The PCR mixture for 16S rRNA gene amplification (50 µl) contained 10 µl 1× GoTag® Flexi green Buffer (Promega), 2.5 µl magnesium chloride (1.25 mM MgCl2, Promega), 1 µl dNTP mix (1.25 mM, Promega), 2.5 µl of each primer (10 µM), 0.25 µl GoTaq® DNA polymerase (100 U, Promega), 3 µl of template DNA and 28.25 µl of miliQ H2O. The PCR was performed under the following conditions: initial denaturation at 95°C for 5 min; followed by 40 cycles at 95°C for 45 s, 60°C for 45 s, 72°C for 1.15 min; and final extension at 72°C for 7 min. For the 18S rRNA gene amplification, the PCR mixture (50 µl) contained 25 µl EmeraldAmpMax PCR Master Mix© (Takara Bio, USA), miliQ H2O (17 µl), 2 µl of each of the primers (10 µM) and 4 µl of template DNA. The PCR reaction was performed under the following conditions: initial denaturation at 98°C for 30 s; followed by 35 cycles at 98°C for 10 s, 55°C for 30 s and 72°C for 1 min; and final extension at 72°C for 10 min. The PCR products were quality-assessed on agarose gels prior to purification with the StartaPrep PCR Purification Kit (Agilent Technologies, Inc.). The purified products were Sanger-sequenced by a commercial provider (Macrogen, the Netherlands).
Sequence processing, multiple sequence alignments and phylogeny
Partial sequences of 16S rRNA and 18S rRNA genes were checked, edited and paired (5’–3’ and 3’–5’ ends) using Sequencher 4.1.4 (Gene Code Corporation, USA). Blast analysis was completed for all sequences with the blast tool available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Sequences of strain PMFPPE4 are deposited in GenBank under accession numbers KU843868.1 for 16S rRNA gene and MH010869.1 for 18S rRNA gene.
In addition to BLAST identification of strain PMFPPE4, we performed small phylogenetic analyses to identify placement and grouping of PMFPPE4 with other Picochlorum strains. A total of 20 sequences for each gene were included in the phylogenetic analyses (Supplementary table 1), with the exception of the 18S rRNA gene Bayesian inference phylogeny which used 16 unique sequences. Four sequences were proven genetically identical (accession numbers: KT860852, KT860853, KT860854 and JQ315636) and therefore omitted from the alignment prior to analysis. The outgroup in the 16S rRNA gene dataset included four sequences belonging to the marine coccoid prasinophytes Pycnococcus provasolii R.R.L. Guillard and Pycnococcus sp., while the outgroup in the 18S rRNA gene dataset included the freshwater auto-sporulating species Marvania coccoides and M. geminata. Multiple sequence alignments were performed in AliView ver. 1.18 using the Muscle algorithm under default parameters (Larsson, Citation2014). The 16S rRNA gene alignment had length of 1322 nucleotides, of which 229 were marked as variable, 182 as parsimony-informative sites and 47 as singleton sites. The 18S rRNA gene alignment had a length of 1816 nucleotides, of which 159 were marked as variable, 89 as parsimony-informative sites and 68 as singleton sites. Alignments were visually inspected, and no sites were manually excluded. Alignments are available at zenodo link: https://zenodo.org/deposit/1186231.
A separate phylogenetic analysis was performed on each gene dataset by first identifying the best model of nucleotide substitution and rate variation across sites using a model-selection routine available in the program IQtree v. 1.5.5, with the specified command -TESTNEWONLY (Nguyen et al., Citation2014). Model selection was completed using the Bayesian information criterion (BIC) which penalizes for the number of parameters and helps avoid overfitting. Phylogenies were reconstructed using Maximum likelihood (ML) and Bayesian inference (BI), in IQtree v. 1.5.5 (Nguyen et al., Citation2014) and MrBayes v. 3.2.6 (Ronquist et al., Citation2012), respectively. Clade support was assessed using IQtree’s UltraFast bootstrap routine (Minh et al., Citation2013) with 1000 pseudoreplicates specified with the -bb 1000 command. Bayesian analyses were carried out with a mixed-model strategy, whereby various variants of the Generalized time-reversible model (GTR) were sampled in proportion to their posterior probability (MrBayes option ‘nst=mix’). Among-site rate variation in MrBayes was accommodated via a Γ distribution with four rate categories (Γ4) and by estimating the proportion of invariant sites (I). Four simultaneous Markov chain Monte Carlo (MCMC) simulations were run, each composed of one cold and three heated chains for a total of 10 million generations with a sampling frequency of one thousand generations. Stationarity and convergence among the MCMC runs were assessed from the MrBayes output (standard deviation of split frequencies and potential scale reduction factor) and by inspecting the posterior distributions in the program Tracer v. 1.6 (Rambaut et al. Citation2007). The first 25% of the sampled posterior distributions were discarded as burn-in. Unique sequences omitted from 18S rRNA gene Bayesian phylogeny were reintroduced into the Newick tree by manually modifying its Newick file.
Pigment analysis
In addition to the cell counts, pigment analysis was performed daily with high-performance liquid chromatography (HPLC) using 1 ml of exponentially growing PMFPPE4 culture filtered through 0.7-μm-pore-size GF/F filters (Whatman, UK), then freshly frozen in liquid nitrogen. Extraction in 4 ml of cold 90% acetone was performed by sonication, and the extract was cleaned by centrifugation. Pigments separation by reversed phase HPLC followed the protocols of Barlow et al. (Citation1997) and Šilović et al. (Citation2011).
Lipid analysis
For lipid class determination, 50 ml of the PMFPPE4 culture in the mid-exponential and stationary growth phases was filtered through precombusted GF/F filters. The filters were stored for 3 days at −80°C before lipid extraction. Particulate lipids were extracted using a modified one-phase solvent mixture of dichloromethane-methanol-water (Bligh & Dyer, Citation1959). N-nonadecanone (KET) was added to each sample as an internal standard to estimate the recoveries in the subsequent steps of the sample analysis. Extracts evaporated to dryness under a nitrogen atmosphere were re-dissolved in 24 µl dichloromethane and lipid classes were determined by thin-layer chromatography-flame ionization detection (TLC-FID; Iatroscan MK-VI, Iatron, Japan). Eighteen lipid classes, constituting total lipids, were separated on Chroma rods SIII and quantified by an external calibration with a standard lipid mixture, with a hydrogen flow of 160 ml min–1 and air flow of 2000 ml min–1. Quantified lipid classes included: hydrocarbons (HC); lipid degradation indices (fatty acid methyl esters (ME), free fatty acids (FFA), alcohols (ALC), 1,3-diacylglycerols (1,3DG), 1,2-diacylglycerols (1,2DG) and monoacylglycerols (MG)); wax and steryl esters (WE/SE, henceforth discussed as SE (WE represent zooplankton storage lipids (Kattner, Citation1989) and should not be present in phytoplankton monocultures); phytoplankton energy reserves (triacylglycerols (TAG)); membrane lipids including three phospholipids (phosphatidylglycerols (PG), phosphatidylethanolamines (PE) and phosphatidylcholines (PC)); three glycolipids (sulfoquinovosyldiacylglycerols (SQDG), monogalactosyldiacylglycerols (MGDG) and digalactosyldiacylglycerols (DGDG)); sterols (ST); and pigments (PIG). For this study, we do not discuss lipid degradation indices or hydrocarbons and the remaining lipid classes represent cell lipids. The standard deviation determined from duplicate runs accounted for 1–8% of the relative abundance of lipid classes. Total lipid concentrations were obtained by summing cell lipid classes. A detailed description of the procedure is published in Gašparović et al. (Citation2015, Citation2017). Total percentages of determined lipid classes within cells were displayed as a pie chart plotted using R software (version 3.4.3) (R Development Core Team, Citation2008) using the ‘ggplot2’ package (Wickham, Citation2011).
Photosynthesis measurement
Measurements of the photosynthetic characteristics of PMFPPE4 were carried out in a laboratory photosynthetron developed by Üveges et al. (Citation2011) using 72 combinations of temperature and light intensity. Temperature varied, in 5°C increments, from 5 to 40°C and light was adjusted to nine light intensities (0; 15; 40; 125; 230; 250; 500; 800 and 1300 µmol photons m–2 s–1). For cultivation and photosynthesis measurements PMFPPE4 cells were maintained in ASN-III medium (Stanier et al., Citation1979). Photosynthesis measurements and carbon uptake calculations were performed as described by Lengyel et al. (Citation2015). Photosynthetic parameters were calculated in the absence of photoinhibition according to Webb et al. (Citation1974) and, if photoinhibition was observed, β (photoinhibition parameter) and the other parameters were calculated according to Platt et al. (Citation1980). Curves (photosynthesis-light intensity (P-I), photosynthesis-temperature (P-T) and Gaussian curves to find temperature optima of photosynthetic parameters) and the calculation of the photosynthetic variables were carried out in GraFit 7.0 software (Leatherbarrow, Citation2009).
Growth experiments
The growth-rate experiment lasted for 20 days throughout which PMFPPE4 was grown in the following conditions: temperature 22 ± 0.5°C; light 30 μmol photons m–2 s–1 with a photoperiod of 16 h of light: 8 h of dark and continuous shaking on an Orbital Shaker OR100 (Cole Parmer, UK) for 12 h during the light period. The starting culture, consisting of ~106 cells ml–1, was established in 200 ml of f/2 medium in Erlenmeyer flasks in triplicate. Cells were counted daily in triplicates using a Birken-Türk haemocytometer. The number of cells ml–1 was then calculated and the standard deviation (SD) was included in generating the growth rate graph, which was plotted using R version 3.4.3 (R Development Core Team, Citation2008) with the ‘ggplot2’ package (Wickham, Citation2011). Net growth rate (k) of PMFPPE4 was calculated using the equation: k = ln(X1−X0)/(T1−T0); X1 = Cell number at the end of exponential phase; X0 = cell number at the beginning of exponential phase; T1 = number of days at the end of exponential phase; T0 = number of days at the beginning of exponential phase.
Additionally, a dark cultivation experiment was performed. The strain was kept in darkness for one month in four replicates, without shaking or any intervention regarding nutrient or oxygen intake, at a stable temperature of 22 ± 0.5°C. Each week, one replicate was examined under light microscope.
Results
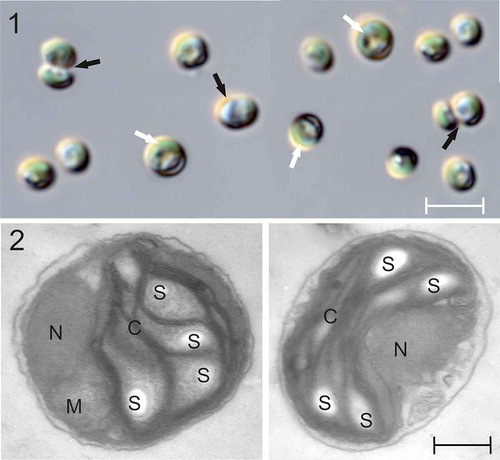
The cells of the isolated algal strain PMFPPE4 are green, spherical to oval, 1.5–3.0 μm (n = 50) in diameter, with a lateral U-shaped chloroplast occupying approximately two-thirds of the cell (). Cells have a smooth surface (, ). No flagella or any other kind of cell appendices were observed. Cells divide by autosporulation into two daughter cells. Neither zoospore formation nor sexual reproduction was observed. TEM revealed that cells contain one nucleus, one mitochondrion, a single, lateral U-shaped chloroplast lacking a pyrenoid, and starch grains are mostly present within the chloroplast (.).
Figs 1–2. Light (LM) and transmission electron (TEM) micrographs of Picochlorum sp. PMFPPE4 strain. Fig. 1: LM micrographs of strain PMFPPE4. White arrows indicate U-shaped chloroplasts and black arrows indicate dividing autospores. Fig. 2: TEM micrographs of strain PMFPPE4. N: nucleus; M: mitochondrion; C: chloroplast; S: starch inclusions. Scale bars: Fig. 1, µm; Fig. 2, 300 nm
The phylogeny of the 16S rRNA gene confirmed placement of the strain PMFPPE4 in the paraphyletic genus Picochlorum (Bayesian posterior probability (BPP)/Bootstrap values (BS), BPP/BS = 1/92 ()), together with other cultured Picochlorum strains without species affiliation, BPP/BS = 0.59/80 (). The most similar strains to PMFPPE4 were all from the Roscoff Culture Collection (RCC), and isolated from the Pacific Ocean (RCC1034, RCC289 and RCC13) and the Mediterranean Sea (RCC9), indicating the cosmopolitan presence of this coccoid pico green alga (). The sister clade containing two additional unknown RCC strains of Picochlorum sp. (RCC846 and RCC945) diverged from the PMFPPE4 clade with great support BPP/BS=1/99 (). The outgroup represented Pycnococcus provasolii and three unidentified Pycnococcus species as root to the tree (BPP/BS=1/100; ).
Fig. 3. Consensus phylograms inferred with Bayesian inference (BI) and Maximum Likelihood (ML) for 16S rRNA (a) and 18S rRNA gene (b). Bayesian posterior probability (BPP) and Maximum likelihood bootstrap values (BS) over 0.5/50 are indicated above branches. All taxa names consist of genus and species name, then strain (if specified in literature)
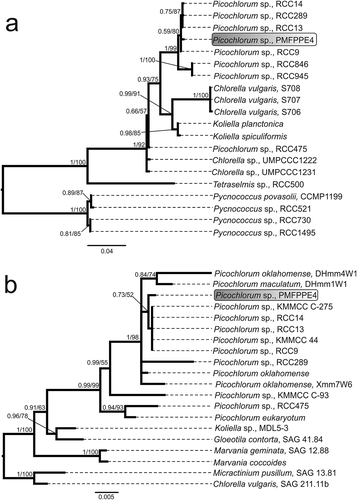
The phylogeny of the 18S rRNA gene also confirmed taxonomic assignation of the strain PMFPPE4 to the monophyletic genus Picochlorum (BPP/BS = 0.99/99, ) by grouping it with five other unknown cultured Picochlorum strains (BPP/BS = 0.73/52; ). Besides the RCC strains (RCC9, 13 and 14), these included strains KMMCC C-275 and KMMCC 44 isolated from the Yellow Sea (). The strain RCC289, Picochlorum oklahomense and P. oklahomense strain Xmm7W6 branched off the PMFPPE4 clade with high support (BPP/BS = 1/98, ). Furthermore, the genus consists of a smaller clade represented by P. oklahomense strain DHmm4W1 and P. maculatum DHmm1W1 (BPP/BS = 0.84/74; ); Picochlorum sp. strain KMMCC C-93 branching off with BPP/BS = 0.99/55; and a clade consisting of Picochlorum sp. RCC475 and P. eukaryotum (BPP/BS = 0.94/93; ). The tree root was comprised of the Micractinium pusillum strain SAG 13.81 and the Chlorella vulgaris strain SAG 211.11b ().
HPLC analyses of pigment content revealed chlorophylls a and b, lutein, β-carotene, violaxanthin and neoxanthin.
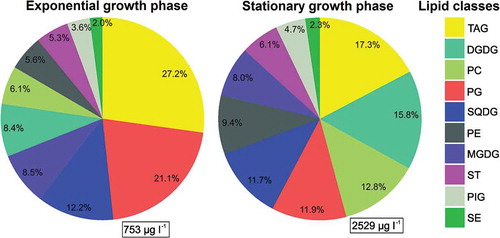
Considerable amounts of particulate lipids were detected in the exponential (day 12) and stationary (day 20) growth phases (753 and 2529 µg l–1 on average; ). The most abundant class was TAG (27.2% and 17.3% in the exponential and stationary growth phase, respectively; ). Regarding membrane-forming lipids, the most abundant were the phospholipids PG (21.1%) and the glycolipids SQDG (12.2%) during the exponential phase, and the stationary phase was characterized by an increased share of the glycolipids DGDG (15.8%) and phospholipids PC (12.8%) ().
Fig. 4. Distribution of Picochlorum sp. PMFPPE4 strain lipid classes during exponential and stationary growth phase. Total lipid concentrations are given in μg l-1 in rectangle (right bottom corner), whereas the relative importance is given in % of total lipids. Abbreviations: TAG: triacylglycerols, DGDG: digalactosyldiacylglycerols, PC: phosphatidylcholines, PG: phosphatidylglycerols, SQDG: sulfoquinovosyldiacylglycerols, PE: phosphatidylethanolamines, MGDG: monogalactosyldiacylglycerols, ST: sterols, PIG: pigments, SE: steryl esters
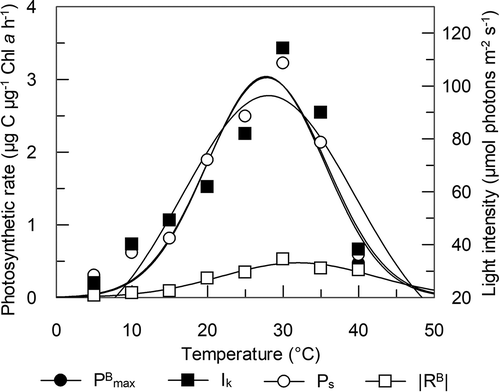
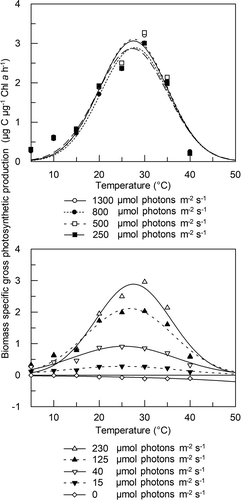
The photosynthesis-light intensity (P-I) curves of PMFPPE4 (Supplementary fig. 1) showed an increase in photosynthetic activity with increasing temperature. The highest value was measured at 30°C. The calculated photosynthetic variables () show temperature dependence. Biomass-specific maximal photosynthetic activity (PBmax) increased with temperature (), the highest value was recorded at 30°C, although the temperature optimum was at 27.7°C (). PBmax increased by an order of magnitude along the temperature gradient: at 5°C PBmax it was 0.304 and at 30°C it was 3.222 µg C µg–1 Chl a h–1. Since photoinhibition was detected only at 40°C, maximal production obtained in the absence of photoinhibition (Ps) differs from PBmax only at the highest temperature. The photoadaptation parameter (Ik) also increased with temperature, and the highest value was calculated at 30°C (114.3 µmol photons m–2 s–1). Good light utilization (α) of the strain was found along the temperature gradient, and highest values were observed at temperature range 20–35°C. Biomass-specific respiration of the strain increased with increasing temperature, reaching a maximum at ~32°C, then slightly decreasing.
Table 1. Photosynthetic parameters of a Picochlorum sp. PMFPPE4 at different temperatures
Fig. 5. Temperature dependence of 4 photosynthetic parameters of Picochlorum sp. PMFPPE4 strain in laboratory experiment. PBmax: Biomass specific maximal photosynthetic production (μg C μg-1 Chl a h-1), Ps: Maximal production obtained in the absence of photoinhibition; without photoinhibition it is equal to PBmax (μg C μg-1 Chl a h-1), Ik: photoadaptation parameter (μmol m-2 s-1), RB: biomass specific respiration (μg C μg-1 Chl a h-1)
The P-T curves showed a similar level of photosynthetic activity in the light intensity range of 230–1300 µmol photons m–2 s–1. Maximum biomass-specific photosynthetic activity varied between 2.89 and 3.10 µg C µg–1 Chl a h–1 (). The temperature optimum for photosynthesis in this light intensity range varied between 27.3 and 27.8°C. At lower light intensities, a remarkable decrease in photosynthetic activity and optimum temperature was observed. At 125 µmol photons m–2 s–1 the maximum photosynthetic activity dropped to 2.1 µg C µg–1 Chl a h–1 and the temperature optimum decreased to just below 27°C. At the two lowest light intensities the decrease was even more pronounced: at 40 and 15 µmol m–2 s–1 the highest photosynthetic activity was at 25.4 and 24.9°C, respectively, with values of 0.92 and 0.28 C µg–1 Chl a h–1, respectively.
Fig. 6. Photosynthesis-temperature (P-T) curves of the Picochlorum sp. PMFPPE4 strain measured in different light intensity ranges
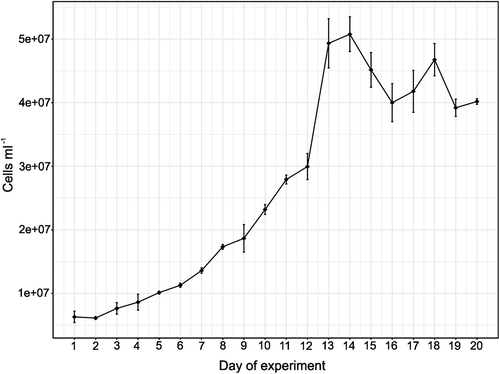
PMFPPE4 cells showed acclimatization and steady growth during the first 7 days of the growth experiment, with an average abundance of 9 × 106 cells ml–1 per day and net growth rate (k) was very high (2.90 day–1). Two weeks after inoculation the culture moved into the stationary growth phase and cells began to aggregate at the bottom of the Erlenmeyer flasks. Cell count stabilized during the last 5 days of the experiment to average values of 4 × 107 ± 2 × 106 cells ml–1 ().
Fig. 7. Line graph of average Picochlorum sp. PMFPPE4 strain cell abundances with standard deviation measured by hemocytometer during over a period of 20 days
Dark cultivation showed that survival of the PMFPPE4 strain was possible without photosynthesis. After a one-month growth period in darkness the strain survived, although cell size was generally smaller (minimum cell diameter was 1.5 µm) in comparison with those grown in 16:8 h light:dark periods.
Discussion
Morphological, ecological and genetic comparisons with similar taxa
Picochlorum sp. PMFPPE4 is morphologically similar to the genus Neocystis Hindák in cell shape and parietal chloroplast, but differs in the number of autospores and the lack of a mother cell wall around vegetative cells (Ettl & Gartner, Citation2014). Similarities to the genus Chlorella and ‘Chlorella-like’ organisms were high, except that species belonging to Chlorella always possess a pyrenoid, which is absent in the genus Picochlorum (Bock et al., Citation2011). There were even greater similarities to the genus Nannochloris Naumann, but without a designated type species, the validity of this genus is questionable (Guiry & Guiry, Citation2019).
To date several species from the genus Nannochloris have been transferred to Picochlorum, indicating there are insufficient distinguishable characters to separate these two genera and leading taxonomists to recognize only the genus Picochlorum (Henley et al., Citation2004; D. Vaulot, personal communication). The lack of distinguishing charateristics and the discussion of Hepperle & Krienitz (Citation2001) regarding difficulties in identification of the so-called ‘Chlorella-’ and ‘Nannochloris-like’ algae, led Henley et al. (Citation2004), using molecular support (18S rDNA phylogeny), to move 13 marine/saline isolates from ‘Nannochloris-like’ algae into Picochlorum gen. nov. W.J.Henley, J.L.Hironaka, L.Guillou, M.A.Buchheim, J.A.Buchheim, M.W.Fawley & K.P.Fawley. At present, the genus Picochlorum comprises five species, three of which are taxonomically accepted: P. oklahomense Hironaka (as a type species), P. maculatum (Butcher) Henley et al. and P. atomus (Butcher) Henley et al. (Guiry & Guiry, Citation2019). There are few marine Trebouxiophyceae genera besides Picochlorum: Chlorella, which is mostly freshwater, but there are few known marine species (1.5–10 µm in diameter); Elliptochloris Tschermak-Woess (5–10 µm in diameter); Carolibrandtia R.Hoshina & T.Nakada (symbiotic green coccoid in ciliates); and Phyllosiphon J.G.Kühn (biofilm-associated siphonous parasitic green algae (Motti et al., Citation2005; Procházková et al., Citation2015; Tragin et al., Citation2016; Hoshina et al., Citation2018), from which the genus Picochlorum differs significantly in cell size, ecology and absence of a pyrenoid. Pseudochloris B.Somogyi, T.Felföldi & L.Vörös is an additional trebouxiophyte genus described by Somogyi et al. (Citation2013), comprising one species: P. wilhelmii, isolated from aquarium seawater. Pseudochloris is morphologically similar to Picochlorum sp. PMFPPE4 in having spherical to oval cells, but the cells seem to be larger (1.6–3.9 µm wide and 1.8–4.0 µm long). The cell ultrastructure of Pseudochloris includes peroxisomes and numerous plastoglobuli, which were not observed in Picochlorum sp. PMFPPE4 (Somogyi et al., Citation2013). Both genera divide by autosporulation (Somogyi et al., Citation2013) and pigment composition is similar – Chl a and b, lutein, violaxanthin and neoxanthin (with the addition of antheraxanthin, zeaxanthin and α-carotene in Pseudochloris). Phylogeny based on the 18S rRNA gene defines P. wilhelmii as sequences belonging to previously misidentified Nannochloris eucaryotum strain UTEX 2505 (JX235961), N. eucaryotum strain SAG 66.87 (JX235962), Chlorella minutissima strain -1 1.9 (X56102) and C. minutissima SAG 1.80 (AB006046) which clearly illustrates the problems with misidentification of Clorella- or Nannochloris-like organisms (Somogyi et al., Citation2013).
Somogyi et al. (Citation2013) represent the genus Picochlorum in their phylogeny as monophyletic, branched off the newly described genus Pseudochloris, therefore acknowledging their genetic diferences. Other classes of algae have also pico-sized non-motile coccoid representatives in the marine environment, which further complicates the identification of Picochlorum. Within the class Mamiellophyceae are genera cosmopolitan and numerically important genera in coastal areas: Bathycoccus W.Eikrem & J.Throndsen and Ostreococcus C.Courties & M.-J.Chrétiennot-Dinet (Moreau et al., Citation2012). Morphological differences to the genus Picochlorum are significant for Bathycoccus, which has specific spider-web-like scales, produced by the Golgi apparatus, on the cell surface (Moreau et al., Citation2012). The smallest known single-celled eukaryotic genus Ostreococcus, species O. tauri has more morphological similarities to Picochlorum sp. PMFPPE4, in terms of simple cell ultrastructure, containing a single chloroplast with one starch granule without pyrenoid and a mitochondrion located between the nucleus and chloroplast (Chrétiennot-Dinet et al., Citation1995). However, pigment composition of O. tauri cells is quite different to Picochlorum sp. PMFPPE4 cells, O. tauri cells contain chl c-like, siphonaxanthin-like 1 and siphonaxanthin-like 2, antheracanthin, zeaxanthin and α-carotene, which are absent in Picochlorum sp. PMFPPE4 (Chrétiennot-Dinet et al., Citation1995). Class Pinguiophyceae contains the marine coccoid genus Pinguiococcus R.A.Andersen, D.Potter, D.& J.C.Bailey comprised of one species, P. pyrenoidosus (collected and described from an aquaculture tank), which also morphologically resembles Picochlorum sp. PMFPPE4 in having spherical to irregular cells with one parietal chloroplast seen in light microscopy (Andersen et al., Citation2002). However, examining cell ultrastructure in TEM, P. pyrenoidosus cells have one stalked pyrenoid in the chloroplast and numerous vacuoles (Andersen et al., Citation2002). Additionally, when observed in growing culture, cells of P. pyrenoidosus have surface protuberances caused by growing vacuoles, and in older cultures cells develop fine extensions of the protoplasm and large storage vacuoles (Andersen et al., Citation2002). Furthermore, the genera Pycnococcus (Pyramimonadophyceae), Prasinoderma (Palmophyllophyceae) and Nannochloro-psis (Eustigmatophyceae) all resemble Picochlorum in LM. Pycnococcus provasolii most closely resembles Picochlorum sp. PMFPPE4 in having spherical, subspherical or ovoid cells ranging from 1.5–4.0 µm in diameter with one cup-shaped chloroplast (Guillard et al., Citation1991). However, the cell ultrastructure observed in TEM differs as one or two pyrenoids are present and the outer mitochondrial membrane protrudes into the chloroplast region of the pyrenoid (Guillard et al., Citation1991). Prasinoderma singularis Jouenne, a solitary coccoid prasinophyte isolated from the South-east Pacific Ocean, can be misidentified in LM as Picochlorum sp. PMFPPE4 owing to cell size (2.2–5.5 µm in diameter) and a single bilobed cupuliform chloroplast. However, the starch-sheath-covered pyrenoid is usually clearly visible in this species, which is a delimiting factor concerning identification of cells belonging to Picochlorum (Jouenne et al., Citation2011). In addition, the pigment of P. singularis differs from Picochlorum sp. PMFPPE4 as it contains Mg-3,8-divinylphaeoporphyrine a5 monomethyl ester, prasinoxanthin, uriolide, micromonol, zeaxanthin and antheraxanthin (Jouenne et al., Citation2011). Finally, cells belonging to Nannochloropsis granulata B.Karlson & D.Potter, isolated and described from the North-east Atlantic Ocean, are commonly mistaken for Picochlorum cells, as in LM they are similar in size (2–4 µm in diameter), chloroplast and absence of a pyrenoid (Karlson et al., Citation1996). However, cells belonging to N. granulata can have two chloroplasts and they always posses up to five refractile granules in the cytoplasm (Karlson et al., Citation1996). N. granulata also has different pigments to Picochlorum sp. PMFPPE4: vaucheriaxanthin like pigment, zeaxanthin and canthaxanthin (Karlson et al., Citation1996).
Examples of defining genera within chlorophytes using multilayer approaches are common (Chrétiennot-Dinet et al., Citation1995; Bock et al., Citation2011 and references therein; de la Vega et al., Citation2011; Somogyi et al., Citation2013; Gonzalez-Esquer et al., Citation2018). Physicochemical characteristics of cultured PPE representatives can help with positioning certain strains, as Dahmen et al. (Citation2014) showed with the identification of Picochlorum sp. strain CTM 20019, or in the examples of Ostreococcus, Pycnococcus, Prasinoderma, Nannochloropsis, etc. discussed above. However, molecular data is the most common tool used to define unknown picoalgal strains such as as Picochlorum (de la Vega et al., Citation2011; Watanabe & Fujii, Citation2016; Gonzalez-Esquer et al., Citation2018) or other genera, such as Pseudochloris (Somogyi et al., Citation2013).
In both phylogenies, PMFPPE4 was most similar to other Picochlorum strains from the RCC, of which none has yet been investigated thoroughly. In the 18S rRNA phylogeny, Picochlorum strains RCC13 (Pacific Ocean), RCC14 (Atlantic Ocean) and RCC9 (Mediterranean Sea) and KMMCC C-275 and KMMCC 44 isolated from the Yellow Sea were genetically identical and probably belong to the same species. PMFPPE4 grouped with this genetically identical clade, suggesting possible conspecificity. However, further investigation would be needed, since conspecificity within the genus Picochlorum has yet to be thoroughly investigated, although it is possible (Krienitz et al. Citation1996, Citation2009; Henley et al. Citation2004). The lack of taxonomic identification within the genus suggests the existence of new and undescribed species and a need for their description in future. However, more extensive morphological effort (thorough TEM examination of all genetically similar strains in vegetative cells and cell divisions) will be needed in order to correctly link those genetically very similar strains. In the 16S rRNA gene phylogeny, not one Picochlorum sequence is identified to species level, and in the 18S rRNA gene phylogeny, only three sequences of taxonomically accepted Picochlorum species are available. The remainder are unknown strains, indicating both poor sequence coverage, lack of taxonomic studies within the genus and usage of gene markers which do not provide sufficient resolution for species delimitation. A possible upgrade in genetic identification of Picochlorum species would be use of another gene marker, such as ITS which already proved to be excellent in delimiting genetically highly similar organisms due to secondary structure comparisons (Škaloud et al., Citation2016; Garcia da Silva et al., Citation2017; Temraleeva & Moslalenko, Citation2019). Under-appreciation of Picochlorum until now is most certainly due to its minute size, the inability to identify cells in environmental samples by their autofluorescence (i.e. flow cytometry), the difficulty in cultivation, and the fact that molecular genetic research on minute coccoid algae is scarce (Barcytė et al., Citation2017). Although potentially undescribed microorganisms can be obtained from current field samples, re-examination of established cultures from public collections such as RCC, NCMA or UTEX is of extreme importance and would be the next step following our study.
Physiology and biotechnological potential of Picochlorum sp. PMFPPE4
Pichochlorum strains are known to grow rapidly (de la Vega et al., Citation2011; Watanabe & Fujii, Citation2016; Gonzalez-Esquer et al., Citation2019). PMFPPE4 entered its exponential phase between days 7–14 and the stationary phase after day 15. In P. oklahomense, maximum biomass concentration was reached at around 18 days, after which it declined (Zhu & Dunford, Citation2013). These results combined illustrate the necessity to harvest biomass of Picochlorum species as soon as the maximum biomass concentration is reached, in the exponential phase.During lipid screening, Picochlorum sp. PMFPPE4 strain was cultivated under nutrient-rich conditions and standard 16:8 light:dark regime. The high TAG level observed, particularly in the exponential phase, was unexpected. TAGs are algal energy-storage lipids. The accumulation of TAG in phytoplankton is usually attributed to nitrogen deprivation, as found in both oligotrophic seas and nitrogen-depleted phytoplankton monocultures (e.g. Parrish & Wangersky, Citation1987; Bourguet et al., Citation2009; Guschina & Harwood, Citation2009; Novak et al., Citation2019). PPEs are a dominant phytoplankton fraction in oligotrophic habitats, as high surface to volume ratio is favourable for nutrient uptake (Marañón, Citation2009). The observed TAG accumulation can be explained as a life strategy to increase surface to volume ratio under conditions of abundant nutrient supply to optimize nutrient uptake (Marañón, Citation2009). We can also hypothesize that accumulation of TAG by PMFPPE4 could indicate an evolutionary preparation by the strain for growth under unfavourable conditions. A similar high-lipid-producing strain of Picochlorum sp. investigated by Tran et al. (Citation2014), and other microalgal strains investigated by Hu et al. (Citation2008), showed higher lipid production as a direct consequence of growth in unfavourable conditions (at 15°C, which was not the photosynthesis optimum for the investigated strain), therefore the results of this study become even more interesting. Furthermore, during the transition from the exponential to stationary growth phase lipid remodelling took place, characterized by increased proportion of membrane-forming DGDG and PC. The glycolipids DGDGs and MGDGs are the major class of lipids in the membranes of plastids, where they are required not only as bulk constituents of photosynthetic membranes but also for the photosynthetic reaction itself (Kobayashi et al., Citation2007). The bilayer-forming DGDG plays an important role in the structural organization of the photosynthetic apparatus (Härtel et al., Citation1997) by stabilizing the thylakoid bilayer structure, while MGDG supports the fluidity of the thylakoid membrane and therefore the velocity of electron flow (Mock & Kroon, Citation2002). We assume that TAGs stored during the exponential phase were used to synthesize DGDG and PC, which are more important for physiological processes and cell integrity, during the stationary growth phase when nutrients are still plentiful in the medium. The phospholipid PC is one of the major structural lipids in the outer membrane, where zwitterionic PC and PE represent up to 68–80% of the structural phospholipids (van Meer et al., Citation2008). Lipid remodelling in the exponential, early stationary and late stationary phases is also observed for diatoms (Su et al., Citation2013). The lipid composition of the PMFPPE4 strain is mostly congruent with other algal species, but it should be noted that lipid composition of plankton cells varies according to environmental factors (Guschina & Harwood, Citation2009). However, the high amount of particulate lipids in cells of Picochlorum sp. PMFPPE4 (among which were TAGs, identified as dominant in cells both in the exponential and stationary growth phases, and phospholipids in membranes during the exponential growth phase) makes this strain exceptional in investigations so far (de la Vega et al., Citation2011; Dahmen et al., Citation2014; Tran et al., Citation2014; Gonzalez-Esquer et al., Citation2019; Dahlin et al., Citation2019; Kumar et al., Citation2019). Therefore, the high TAG content and high growth rate suggest PMFPPE4 could be of interest for biofuel production and growth in outdoor systems. Dahmen et al. (Citation2014) emphasized the biotechnological potential of the genus Picochlorum and demonstrated the feasibility of using a wild Picochlorum sp. as feedstock for aquaculture, human nutrition or biodiesel production. Additionally, de la Vega et al. (Citation2011) demonstrated the great potential for Picochlorum sp. HM1 to grow and thrive under adverse conditions, making it a good candidate for outdoor cultivation. Picochlorum sp. ‘soloecismus’ was also noted as a resilient strain with great potential as a platform for production of biofuels and bioproducts (Gonzalez-Esquer et al., Citation2019). Another Picochlorum strain, the genome sequenced Picochlorum sp. ‘renovo’, proved to be a high-growth, halophilic, thermotolerant strain and was also suggested for outdoor growth systems and biofuel production (Dahlin et al., Citation2019).
Increasing photosynthetic activity of Picochlorum strain PMFPPE4 with temperature is consistent with results from research on sister algal genera, such as Chlorella (Yun & Park, Citation2003; Lee et al., Citation2018) and also with other species from different phyla, e.g. Bacillariophyta, Cyanobacteria, Rhodophyta (Coles & Jones, Citation2000; Üveges et al., Citation2012; Lengyel et al., Citation2015; Pálmai et al., Citation2018). The photosynthetic activity along temperature gradient of Picochlorum sp. PMFPPE4 is well below the values for some river cyanobacteria species (Coles & Jones, Citation2000) and the dominant species in tropical alkaline saline lakes (Schagerl et al., Citation2015), but is similar to some dominant saline diatom species (Lengyel et al., Citation2015). The photosynthetic temperature optimum for Picochlorum sp. PMFPPE4 was measured at exactly 27.7°C, which is congruent with that of P. maculatum, which had maximum nutrients consumption at 28°C with a photoperiod of 18h light and 6h dark and 150 µmol photons m–2 s–1 (Kumar et al., Citation2019). Other Picochlorum strains showed optimum growth rates (although photosynthetic activity was not measured) at similar temperatures (25–30°C), and at higher temperatures growth rate declined (de la Vega et al., Citation2011; Foflonker et al., Citation2016). The potential mixotrophy of the species could be a reason for the moderate photosynthetic activity under unfavourable environmental conditions (e.g. low temperature). Green algae typically prefer higher light intensities (~80–500 µmol photons m–2 s–1; Reynolds, Citation1988; Padisák, Citation2004) than our findings, but there are also species with a preference for shade (e.g. Picocystis salinarum; Roesler et al., Citation2002). The low light intensity preference along the temperature gradient, with good light utilization, suggests PMFPPE4 can tolerate or even prefer a light-limited habitat. Conversely, the higher DGDG:MGDG ratio found in our strain indicates it can grow under higher light conditions (Mock & Kroon, Citation2002). Other Picochlorum strains, such as Picochlorum sp. HM, Picochlorum sp. ‘soloecismus’ and Picochlorum sp. ‘renovo’ also show tolerance of a wide range of light intensities (de la Vega et al. Citation2011; Dahlin et al. Citation2019; Gonzalez-Esquer et al. Citation2019). Our findings, including survival of Picochlorum sp. PMFPPE4 in dark conditions for an extended period, further suggest that the ecological niche of this picoalga is wide, in regard to both light regimes and temperature, and that it stores sufficient TAGs in reserves to survive unfavourable conditions. Additionally, a record of PPEs below the photic zone (Babić et al., Citation2017) during the BIOTA cruise suggest a possible switch of Picochlorum sp. PMFPPE4 to a mixotrophic lifestyle in an unfavourable growth environment. The accumulation of PPEs at 280 m depth was not significantly correlated with any environmental variables (e.g. salinity, nutrients or temperature) suggesting that they were found outside their ecological optima (Babić et al., Citation2017). Vertical density gradients were relatively strong in that area and geostrophic currents indicated a strong vertical shear (Babić et al., Citation2017). The shear may have caused vertical instabilities and transport of water parcels from surface to deep layers, which could have been responsible for the occurrence of the maximum of PPEs at depths below the euphotic zone. Selective pressures on preserving photoautotrophic machinery can be relaxed under certain conditions, such as when the energy costs of maintaining the photosynthetic apparatus outweigh the benefits of its products. Picoeukaryotes can use phagocytosis in cases of mixotrophy/heterotrophy (Massana & Logares, Citation2013) and additionally, mixotrophy can be achieved through osmotrophy (the uptake of dissolved organic substrates; Pringsheim, Citation1963; Glibert & Legrand, Citation2006). Given the general morphology of the Picochlorum sp. PMFPPE4 strain (no observed flagella or apparatus for catching prey) and that there is no record of toxin production for prey immobilization and/or disability in the available literature, we can assume that in unfavourable conditions this organism acts as an osmotroph.
This study identified a pico green alga isolated from the south-eastern Adriatic Sea, strain PMFPPE4, as member of the genus Picochlorum, a largely disregarded but widespread and molecularly diversified genus of Trebouxiophyceae. Identification of small coccoid algae can be achieved through a multilayer approach, considering morphology, phylogeny and physiology. The ecological preferences of Picochlorum sp. PMFPPE4 are broad: from shaded (and completely dark) and cooler marine environments (5–10°C), where it may act as an osmotroph; to higher light intensity (114.3 µmol photons m–2 s–1) and temperature (30–40°C), where it has photosynthesis activity maxima and finally photoinhibition (> 40°C). The low light intensity preference along a temperature gradient, with good light utilization, suggests this strain can tolerate a light-limited habitat. In regard to lipids, the most important finding was the ability of the strain to synthesize large amounts of triacylglycerids, which are important for biofuel feedstock and the food industry, and its high DGDG/MGDG ratio which indicates it can tolerate high light conditions. This exceptional strain can also serve as a model green algae in diverse ecological experiments, due to its longevity in cultured conditions.
Supplemetary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at 10.1080/09670262.2020.1757763
Supplementary table 1: List of all taxa with strain information and GenBank accession numbers used for phylogeny inference in this study
Supplementary fig. 1: Consensus phylograms inferred with Bayesian inference (BI) and Maximum likelihood (ML) for 16S rRNA (a) and 18S rRNA gene (b). Bayesian posterior probability (PP) and Maximum likelihood bootstrap values (BS) over 0.5/50 are indicated above branches. All taxa names consist of genus and species name, then strain (if specified in literature).
Author contributions
M. Mucko: original concept, writing and editing manuscript, morphological analyses, culture establishment and experiments and phylogeny analyses and plotting; J. Padisák: photosynthetic activity measurements, writing and editing manuscript; M. Gligora Udovič: photosynthetic activity measurements, writing and editing manuscript; T. Pálmai: photosynthetic activity measurements and plotting, writing and editing manuscript; T. Novak: lipid analysis, writing and editing manuscript; N. Medić: lipid analysis, writing and editing manuscript; B. Gašparović: lipid analysis, writing and editing manuscript; P. Peharec Štefanić: sample preparation and examination for transmission electron microscopy, editing manuscript; S. Orlić: DNA isolation, sequence processing and phylogeny; Z. Ljubešić: original concept, writing and editing manuscript.
TEJP-2019-0136-File008.doc
Download MS Word (116 KB)Acknowledgements
The authors would like to express their thanks to Margie Cepon for critical reading of the manuscript and language corrections and to the crew of research vessel Naše more for their help during the fieldwork. M.M. is thankful to Tanja Vojvoda Zeljko for help during the molecular laboratory work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2020.1757763.
Additional information
Funding
References
- Andersen, R.A., Pottert, D. & Bailey, J.C. (2002). Pinguiococcus pyrenoidosus gen. et sp. nov. (Pinguiophyceae), a new marine coccoid alga. Phycological Research, 50: 57–65.
- Babić, I., Petrić, I., Bosak, S., Mihanović, H., Dupčić Radić, I. & Ljubešić, Z. (2017). Distribution and diversity of marine picocyanobacteria community: targeting of Prochlorococcus ecotypes in winter conditions (southern Adriatic Sea). Marine Genomics, 36: 3–11.
- Barcytė, D., Hodač, L., & Nedbalová, L. (2017). Lunachloris lukesovae gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel coccoid green alga isolated from soil in South Bohemia, Czech Republic. European Journal of Phycology, 52: 281–291.
- Barlow, R.G., Mantoura, R.F.C., Cummings, D.G. & Fileman, T.W. (1997). Pigment chemotaxonomic distributions of phytoplankton during summer in the western Mediterranean. Deep Sea Research Part 2, 44: 833–850.
- Bligh, E.G. & Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 37: 911–917.
- Bock, C., Krienitz, L. & Pröschold, T. (2011). Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea, 11: 293–312.
- Bork, P., Bowler, C., de Vargas, C., Gorsky, G., Karsenti, E. & Wincker, P. (2015). Tara Oceans studies plankton at planetary scale. Introduction. Science 348(6237): 873.
- Bosak, S., Bošnjak, I., Cetinić, I., Mejdandžić, M. & Ljubešić, Z. (2016). Diatom community in the depths of the South Adriatic: an injection of carbon by biological pump. In 41st CIESM Congress.
- Bourguet, N., Goutx, M., Ghiglione, J.F., Pujo-Pay, M., Mevel, G., Momzikoff, A., Mousseau, L., Guigue, C., Garcia, N., Raimbault, P., Pete, R., Oriol, L. & Lefevre, D. (2009). Lipid biomarkers and bacterial lipase activities as indicators of organic matter and bacterial dynamics in contrasted regimes at the DYFAMED site, NW Mediterranean. Deep-Sea Research Part II, 56: 1454–1469.
- Chrétiennot-Dinet, M.J., Courties, C., Vaquer, A., Neveux, J., Claustre, H., Lautier, J. & Machado, M.C. (1995). A new marine picoeucaryote: Ostreococcus tauri gen. et sp. nov. (Chlorophyta, Prasinophyceae). Phycologia, 34: 285–292.
- Claustre, H., Sciandra, A. & Vaulot, D. (2008). Introduction to the special section bio-optical and biogeochemical conditions in the South East Pacific in late 2004: the BIOSOPE program. Biogeosciences Discussion, 5: 605–640.
- Coles, J.F. & Jones, R.C. (2000). Effect of temperature on photosynthesis-light response and growth of four phytoplankton species isolated from a tidal freshwater river. Journal of Phycology, 36: 7–16.
- Dahlin, L.R., Gerritsen, A.T., Henard, C.A., Van Wychen, S., Linger, J.G., Kunde, Y., Hovde, B.T., Starkenburg, S.R., Posewitz, M.C. & Guarnieri, M.T. (2019). Development of a high-productivity, halophilic, thermotolerant microalga Picochlorum renovo. Communications Biology, 2: 1–9.
- Dahmen, I., Chtourou, H., Jebali, A., Daassi, D., Karray, F., Hassairi, I., Sayadi, S., Abdelkafi, S. & Dhouib, A. (2014). Optimisation of the critical medium components for better growth of Picochlorum sp. and the role of stressful environments for higher lipid production. Journal of the Science of Food and Agriculture, 94: 1628–1638.
- de la Vega, M., Diaz, E., Vila, M. & León, R. (2011). Isolation of a new strain of Picochlorum sp. and characterization of its potential biotechnological applications. Biotechnology Progress, 27: 1535–1543.
- de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahé, F., Logares, R., Lara, E., Berney, C., Le Bescot, N., Probert, I., Carmichael, M., Poulain, J., Romac, S., Colin, S., Aury, J.M., Bittner, L., Chaffron, S., Dunthorn, M., Engelen, S., Flegontova, O., Guidi, L., Horák, A., Jaillon, O., Lima-Mendez, G., Lukeš, J., Malviya, S., Morard, R., Mulot, M., Scalco, E., Siano, R., Vincent, F., Zingone, A., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Tara Oceans Coordinators, Acinas, S.G., Bork, P., Bowler, C., Gorsky, G., Grimsley, N., Hingamp, P., Iudicone, D., Not, F., Ogata, H., Pesant, S., Raes, J., Sieracki, M.E., Speich, S., Stemmann, L., Sunagawa, S., Weissenbach, J., Wincker, P. & Karsenti, E. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science, 348: 1261605.
- Decelle, J., Romac, S., Stern, R.F., Bendif, E.M., Zingone, A., Audic, S., Guiry, M.D., Guillou, L., Tessier, D., Le Gall, F., Gourvill, P., Dos Santos, A.L., Probert, I., Vaulot, D., de Vargas, C. & Christen, R. (2015). PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Molecular Ecology Resources, 15: 1435–1445.
- Dı́ez, B., Pedrós-Alió, C. & Massana, R. (2001). Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Applied and Environmental Microbiology, 67: 2932–2941.
- Duarte, C.M. (2015). Seafaring in the 21st century: the Malaspina 2010 Circumnavigation Expedition. Limnology and Oceanography Bulletin, 24: 11–14.
- Ettl, H. & Gärtner, G. (2014). Syllabus der boden-, luft-und flechtenalgen. Springer-Verlag, Stuttgart.
- Foflonker, F., Ananyev, G., Qiu, H., Morrison, A., Palenik, B., Dismukes, G.C. & Bhattacharya, D. (2016). The unexpected extremophile: tolerance to fluctuating salinity in the green alga Picochlorum. Algal Research, 16: 465–472.
- Fuller, N.J., Campbell, C., Allen, D.J., Pitt, F.D., Le Gall, F., Vaulot, D. & Scanlan, D.J. (2006a). Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquatic Microbial Ecology, 43: 79–93.
- Fuller, N.J., Tarran, G., Cummings, D.G., Woodward, M.S., Orcutt, K.M., Yallop, M., Le Gall, F. & Scanlan, D.J. (2006b). Molecular analysis of photosynthetic picoeukaryote community structure along an Arabian Sea transect. Limnology and Oceanography, 51: 2052–2514.
- Galloway, A.W. & Winder, M. (2015). Partitioning the relative importance of phylogeny and environmental conditions on phytoplankton fatty acids. PLoS ONE, 10: e0130053.
- Garcia da Silva, T., Bock, C., Sant’Anna, C.L., Bagatini, I.L., Wodniok, S. & Vieira, A. A.H. (2017). Selenastraceae (Sphaeropleales, Chlorophyceae): rbcL, 18S rDNA and ITS-2 secondary structure enlightens traditional taxonomy, with description of two new genera, Messastrum gen. nov. and Curvastrum gen. nov. Fottea, 17: 1–19.
- Gašparović, B., Kazazić, S.P., Cvitešić, A., Penezić, A. & Frka, S. (2015). Improved separation and analysis of glycolipids by Iatroscan thin-layer chromatography–flame ionization detection. Journal of Chromatography A, 1409: 259–267.
- Gašparović, B., Kazazić, S.P., Cvitešić, A., Penezić, A. & Frka, S. (2017). Corrigendum to “Improved separation and analysis of glycolipids by Iatroscan thin-layer chromatography–flame ionization detection” [Journal of Chromatography A. 1409 (2015): 259–267]. Journal of Chromatography A, 1521: 168–169.
- Glibert, P.M. & Legrand, C. (2006). The diverse nutrient strategies of harmful algae: focus on osmotrophy. In The Ecology of Harmful Algae (Grane´li, E. & Turner, J., editors), 163–175. Springer-Verlag, New York.
- Gonzalez-Esquer, C.R., Twary, S.N., Hovde, B.T. & Starkenburg, S.R. (2018). Nuclear, chloroplast, and mitochondrial genome sequences of the prospective microalgal biofuel strain Picochlorum soloecismus. Genome Announcments, 6: e01498–17.
- Gonzalez-Esquer, C.R., Wright, K.T., Sudasinghe, N., Carr, C.K., Sanders, C.K., Turmo, A., Kerfeld, C.A., Twary, S. & Dale, T. (2019). Demonstration of the potential of Picochlorum soloecismus as a microalgal platform for the production of renewable fuels. Algal Research, 43: 101658.
- Grob, C., Ulloa, O., Claustre, H., Huot, Y., Alarcon, G. & Marie, D. (2007). Contribution of picoplankton to the total particulate organic carbon concentration in the eastern South Pacific. Biogeosciences, 4: 837–852.
- Guillard, R.R., Keller, M.D., O’Kelly, C.J. & Floyd, G.L. (1991). Pycnococcus provasolii gen. et sp. nov., a coccoid prasinoxanthin-containing phytoplankter from the western North Atlantic and Gulf of Mexico. Journal of Phycology, 27: 39–47.
- Guillou, L., Chrétiennot-Dinet, M.J., Medlin, L.K., Claustre, H., Loiseaux-de Goër, S. & Vaulot, D. (1999). Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta). Journal of Phycology, 35: 368–381.
- Guiry, M.D. & Guiry, G.M. (2019). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Available from: http://www.algaebase.org.
- Guschina, I.A. & Harwood, J.L. (2009). Algal lipids and effect of the environment on their biochemistry. In Lipids in Aquatic Ecosystems ( Kainz, M., Brett, M. & Arts, M., editors), 1–24. Springer, New York.
- Härtel, H., Lokstein, H., Dörmann, P., Grimm, B. & Benning, C. (1997). Changes in the composition of the photosynthetic apparatus in the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiology, 115: 1175–1184.
- Henley, W.J., Hironaka, J.L., Guillou, L., Buchheim, M.A., Buchheim, J.A., Fawley, M.W. & Fawley, K.P. (2004). Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia, 43: 641–652.
- Hepperle, D. & Krienitz, L. (2001). Systematics and ecology of chlorophyte picoplankton in German inland waters along a nutrient gradient. International Review of Hydrobiology, 86: 269–284.
- Hoshina, R., Kobayashi, M., Suzaki, T. & Kusuoka, Y. (2018). Brandtia ciliaticola gen. et sp. nov. (Chlorellaceae, Trebouxiophyceae) a common symbiotic green coccoid of various ciliate species. Phycological Research, 66: 76–81.
- http://sagdb.uni-goettingen.de/; accessed 1.03.2018. at 16:00.
- https://ncma.bigelow.org/; accessed 1.03.2018. at 16:00.
- https://utex.org/; accessed 1.03.2018. at 16:00.
- Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M. & Darzins, A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant Journal, 54: 621–639.
- http://roscoff-culture-collection.org/
- Jouenne, F., Eikrem, W., Le Gall, F., Marie, D., Johnsen, G. & Vaulot, D. (2011). Prasinoderma singularis sp. nov. (Prasinophyceae, Chlorophyta), a solitary coccoid prasinophyte from the South-East Pacific Ocean. Protist, 162: 70–84.
- Karlson, B., Potter, D., Kuylenstierna, M. & Andersen, R.A. (1996). Ultrastructure, pigment composition, and 18S rRNA gene sequence for Nannochloropsis granulata sp. nov. (Monodopsidaceae, Eustigmatophyceae), a marine ultraplankter isolated from the Skagerrak, northeast Atlantic Ocean. Phycologia, 35: 253–260.
- Kattner, G. (1989). Lipid composition of Calanus finmarchicus from the North Sea and the Arctic. A comparative study. Comparative Biochemistry and Physiology, 94: 185–188.
- Kirk, J.T.O. (1994). Light and Photosynthesis in Aquatic Ecosystems. Cambridge University Press, Cambridge.
- Knight-Jones, E.W. (1951). Preliminary studies of nanoplankton and ultraplankton systematics and abundance by a quantitative culture method. Journal du Conseil, 17: 140–155.
- Kobayashi, K., Kondo, M., Fukuda, H. & Nishimura, M. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proceedings of the National Academy of Sciences USA, 104: 17216–17221.
- Krienitz, L., Bock, C., Dadheech, P.K. & Pröschold, T. (2011). Taxonomic reassessment of the genus Mychonastes (Chlorophyceae, Chlorophyta) including the description of eight new species. Phycologia, 50: 89–106.
- Krienitz, L., Huss, V.A.R. & Hümmer, C. (1996). Picoplanktonic Choricystis species (Chlorococcales, Chlorophyta) and problems surrounding the morphologically similar ‘Nannochloris-like algae’. Phycologia, 35: 332–341.
- Kumar, S.D., Ananth, S., Santhanam, P., Ahamed, A.P. & Thajuddin, N. (2019). Effect of photoperiod (PP) and photosynthetic photon flux intensity (PPFI) on nutrients consumption, growth and lipid profile of unusual microalga Picochlorum maculatum (PSDK01) in shrimp culture effluent. Indian Journal of Experimental Biology, 57: 105–115.
- Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics, 30: 3276–3278.
- Leatherbarrow, R. (2009). GraFit data analysis software for Windows. 7.0.3 edn. Erithacus Software Ltd. Horley.
- Lee, K.K., Lim, P.E., Poong, S.W., Wong, C.Y., Phang, S.M. & Beardall, J. (2018). Growth and photosynthesis of Chlorella strains from polar, temperate and tropical freshwater environments under temperature stress. Chinese Journal of Oceanology and Limnology, 36: 1266–1279.
- Lengyel, E., Kovács, A.W., Padisák, J. & Stenger-Kovács, C. (2015). Photosynthetic characteristics of the benthic diatom species Nitzschia frustulum (Kützing) Grunow isolated from a soda pan along temperature-, sulfate- and chloride gradients. Aquatic Ecology, 49: 401–416.
- Lepère, C., Demura, M., Kawachi, M., Romac, S., Probert, I. & Vaulot, D. (2011). Whole-genome amplification (WGA) of marine photosynthetic eukaryote populations. FEMS Microbiology Ecology, 76: 513–523.
- Li, W.K.W. (1994). Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnology and Oceanography, 39: 169–175.
- Liu, J., Huang, J., Sun, Z., Zhong, Y., Jiang, Y. & Chen, F. (2011). Differential lipid and fatty acid profiles of photoautotrophic and heterotrophic Chlorella zofingiensis: assessment of algal oils for biodiesel production. Bioresource Technology, 102: 106–110.
- Marañón, E. (2009). Phytoplankton size structure. In Encyclopedia of Ocean Sciences. 2nd ed. (Steele, J.H., editor), 445–452. Academic Press, Elsevier.
- Massana, R., Balagué, V., Guillou, L. & Pedrós-Alió, C. (2004). Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiology Ecology, 50: 231–243.
- Massana, R. & Logares, R. (2013). Eukaryotic versus prokaryotic marine picoplankton ecology. Environmental Microbiology, 15: 1254–1261.
- Minh, B.Q., Nguyen, M.A.T. & von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30: 1188–1195.
- Mock, T. & Kroon, B.M. (2002). Photosynthetic energy conversion under extreme conditions – II: the significance of lipids under light limited growth in Antarctic sea ice diatoms. Phytochemistry, 61: 53–60.
- Moreau, H., Verhelst, B., Couloux, A., Derelle, E., Rombauts, S., Grimsley, N., Van Bel, M., Poulain, J., Katinka, M., Hofmann-Marriott, M.F., Piganeau, G., Rouzé, P., Da Silva, C., Wincker, P., Van de Peer, Y. & Vandepoele, K. (2012). Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biology, 13: R74.
- Motti, C., Curiel, D., Rismondo, A., Bellemo, G., Dri, C., Checchin, E. & Marzocchi, M. (2005). First report of a species of Prasiola (Chlorophyta: Prasiolacea) from the Mediterranean Sea (Lagoon of Venice). Scientia Marina, 69: 343–346.
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32: 268–274.
- Novak, T., Godrijan, J., Marić Pfannkuchen, D., Djakovac, T., Medić, N., Ivančić, I., Mlakar, M. & Gašparović, B. (2019). Global warming and oligotrophication lead to increased lipid production in marine phytoplankton. Science of the Total Environment, 668: 171–183.
- Padisák, J. (2004). Phytoplankton. In The Lakes Handbook, vol 1. Limnology and Limnotic Ecology (O’Sullivan, P. & Reynolds, C.S., editors), 251–309. Blackwell Science, Oxford.
- Pálmai, T., Szabó, B., Hubai, K.E. & Padisák, J. (2018). Photosynthetic performance of two freshwater red algal species. Acta Botanica Croatica, 77: 135–40.
- Parrish, C.C. & Wangersky P.J. (1987). Particulate and dissolved lipid classes in cultures of Phaeodactylum tricornutum grown in cage culture turbidostats with a range of nitrogen supply rates. Marine Ecology Progress Series 35: 119–128.
- Platt, T., Gallegos, C.L. & Harrison, W.G. (1980). Photoinibition of photosynthesis in natural assemblages of marine phytoplankton. Journal of Marine Research, Instituto Del Mar Del Peru Boletin, Volumen extraordinario.
- Pringsheim, E.G. (1963). Farblose Algen, ein beitrag zur evolutionsforschung. G. Fischer, Stuttgart. Germany.
- Procházková, K., Nemcová, Y., Kulichová, J. & Neustupa, J. (2015). Morphology and phylogeny of parasitic and free-living members of the genus Phyllosiphon (Trebouxiophyceae, Chlorophyta). Nova Hedwigia, 101: 501–518.
- R Development Core Team (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org.
- Rambaut, A., Drummond, A.J. & Suchard, M. (2007). Tracer v1. 6. http://beast.bio.ed.ac.uk.Tracer
- Reynolds, C. (1988). Functional morphology and the adaptive strategies of freshwater phytoplankton. In Growth and Reproductive Strategies of Freshwater Phytoplankton, 388–433. Cambridge University Press, Cambridge.
- Reynolds, E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. Journal of Cell Biology, 17: 208.
- Richardson, T.L. & Jackson, G.A. (2007). Small phytoplankton and carbon export from the surface ocean. Science, 315: 838–840.
- Roesler, C.S., Culbertson, C.W., Etheridge, S.M., Goericke, R., Kiene, R.P., Miller, L.G. & Oremland, R.S. (2002). Distribution, production, and ecophysiology of Picocystis strain ML in Mono Lake, California. Limnology and Oceanography, 47: 440–452.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Schagerl, M., Burian, A., Gruber-Dorninger, M., Oduor, S.O. & Kaggwa, M.N. (2015). Algal communities of Kenyan soda lakes with a special focus on Arthrospira fusiformis. Fottea, 15: 245–257.
- Sieracki, M.E., Poulton, N.J., Jaillon, O., Wincker, P., de Vargas, C., Rubinat-Ripoll, L., Tepanauskas, R., Logares, R. & Massana, R. (2019). Single cell genomics yields a wide diversity of small planktonic protists across major ocean ecosystems. Scientific Reports, 9: 6025.
- Šilović, T., Ljubešić, Z., Mihanović, H., Olujić, G., Terzić, S., Jakšić, Ž. & Viličić, D. (2011). Picoplankton composition related to thermohaline circulation: the Albanian boundary zone (southern Adriatic) in late spring. Estuarine, Coastal and Shelf Science, 91: 519–525.
- Škaloud, P., Friedl, T., Hallmann, C., Beck, A. & Dal Grande, F. (2016). Taxonomic revision and species delimitation of coccoid green algae currently assigned to the genus Dictyochloropsis (Trebouxiophyceae, Chlorophyta). Journal of Phycology, 52: 599–617.
- Somogyi, B., Felföldi, T., Solymosi, K., Flieger, K., Márialigeti, K., Böddi, B. & Vörös, L. (2013). One step closer to eliminating the nomenclatural problems of minute coccoid green algae: Pseudochloris wilhelmii, gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). European Journal of Phycology, 48: 427–436.
- Stanier, R.Y., Deruelles, J., Rippka, R., Herdman, M. & Waterbury, J.B. (1979). Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Microbiology, 111: 1–6.
- Su, X., Xu, J., Yan, X., Zhao, P., Chen, J., Zhou, C., Zhao, F. & Li, S. (2013). Lipidomic changes during different growth stages of Nitzschia closterium f. minutissima. Metabolomics 9: 300–310.
- Temraleeva, A.D. & Moslalenko, S.V. (2019). Application of morphological and molecular systematics for identification of green microalgae of the genus Chlorococcum and some closely related taxa. Microbiology, 88: 27–38.
- Throndsen, J. (1978). The dilution-culture method. In Phytoplankton Manual, 218–224. Unesco, Paris.
- Tragin, M., Lopes dos Santos, A., Christen, R. & Vaulot, D. (2016). Diversity and ecology of green microalgae in marine systems: an overview based on 18S rRNA gene sequences. Perspectives in Phycology, 3: 141–154.
- Tragin, M. & Vaulot, D. (2018). Green microalgae in marine coastal waters: The Ocean Sampling Day (OSD) dataset. Scientific Reports, 8: 14020.
- Tran, D., Giordano, M., Louime, C., Tran, N., Vo, T., Nguyen, D. & Hoang, T. (2014). An isolated Picochlorum species for aquaculture, food, and biofuel. North American Journal of Aquaculture, 76: 305–311.
- Üveges, V., Vörös, L., Padisák, J. & Kovács, A.W. (2011). Primary production of epipsammic algal communities in Lake Balaton (Hungary). Hydrobiologia, 660: 17–27.
- Üveges, V., Tapolczai, K., Krienitz, L. & Padisák, J. (2012). Photosynthetic characteristics and physiological plasticity of an Aphanizomenon flos-aquae (Cyanobacteria, Nostocaceae) winter bloom in a deep oligo-mesotrophic lake (Lake Stechlin, Germany). Hydrobiologia, 698: 263–272.
- van Meer, G., Voelker, D.R. & Feigenson, G.W. (2008). Membrane lipids: where they are and how they behave. Nature Reviews Molecular Cell Biology, 9: 112–124.
- Vaulot, D., Eikrem, W., Viprey, M. & Moreau, H. (2008). The diversity of small eukaryotic phytoplankton (≤ 3 μm) in marine ecosystems. FEMS Microbiology Reviews, 32: 795–820.
- Watanabe, K. & Fujii, K. (2016). Isolation of high-level-CO2-preferring Picochlorum sp. strains and their biotechnological potential. Algal Research, 18: 135–143.
- Webb, W.L., Newton, M. & Starr, D. (1974). Carbon dioxide exchange of Alnus rubra – a mathematical model. Oecologia, 17: 281–291.
- West, N.J., Schönhuber, W.A., Fuller, N.J., Amann, R.I., Rippka, R., Post, A.F. & Scanlan, D.J. (2001). Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology, 147: 1731–1744.
- Wickham, H. (2011). ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics, 3: 180–185.
- Yun, Y.S. & Park, J.M. (2003). Kinetic modeling of the light‐dependent photosynthetic activity of the green microalga Chlorella vulgaris. Biotechnology and Bioengineering, 83: 303–311.
- Zhu, Y. & Dunford, N.T. (2013). Growth and biomass characteristics of Picochlorum oklahomensis and Nannochloropsis oculata. Journal of the American Oil Chemists’ Society, 90: 841–849.
