ABSTRACT
Characiopsis, established by Borzì in 1895, is the largest genus traditionally classified in the class Xanthophyceae. However, Characiopsis-like algae studied over the last five decades using transmission electron microscopy and molecular phylogenetics have all proved to belong to a different class, the Eustigmatophyceae. Despite this, Characiopsis is still treated as a xanthophyte taxon by most algal taxonomy resources, partly because of uncertainties concerning the identity of the type of the genus. Here we document the morphology of 20 morphologically diverse and (mostly) previously unstudied Characiopsis isolates, and establish their phylogenetic position using 18S rRNA and rbcL gene sequence data. We show that these algae all belong to a single clade within the eustigmatophyte subgroup referred to as the Eustigmataceae group. From careful re-examination of previous taxonomic accounts concerning the genus Characiopsis we conclude that its type is undoubtedly Characiopsis minuta (Braun) Borzì (basionym Characium minutum Braun). In view of the loss of the holotype of this species, we designate a neotype, and also a supporting epitype (a cryopreserved culture of one of the studied strains). Our results convincingly show that Characiopsis must be transferred from the Xanthophyceae to the Eustigmatophyceae. Its assignment to the Eustigmataceae group is consistent with our observation of a pyrenoid in most of the strains studied, which distinguishes these algae from pyrenoid-free species previously classified in the genus Characiopsis but recently accommodated in the newly erected genera Neomonodus, Characiopsiella and Munda in the eustigmatophyte family Neomonodaceae. We additionally confirm the previous suggestion that C. minuta is closely related to, if not conspecific with, Pseudocharaciopsis texensis K.W.Lee & Bold, the type of the genus Pseudocharaciopsis, which is thus rendered a junior synonym of Characiopsis. Altogether, our work significantly improves the classification of a charismatic yet poorly known group of algae.
Introduction
The current concept of the class Xanthophyceae (yellow-green algae), one of the most prominent lineages of ochrophyte algae, is largely inherited from the pre-molecular era of phycology. According to the most recent general treatment of the class by Maistro et al. (Citation2017), it comprises about 600 described species in over 90 genera. However, as they emphasize, the traditional classification into orders, families and genera is not congruent with insights from molecular phylogenetics, and a critical revision of xanthophyte systematics is needed. The problem, however, is not only the inaccurate internal classification of the group, but also that the Xanthophyceae as presently circumscribed is polyphyletic, including organisms that are not directly related to the ‘core’ of the class. Various traditional xanthophytes, as presented in the most recent monographic account (Ettl, Citation1978), were later shown to be representatives of unrelated groups, such as Chlorarachniophyta (Hibberd & Norris, Citation1984) and green algae (Gärtner & Schragl, Citation1988; Darienko et al., Citation2010; Eliáš et al., Citation2013). Other taxa may still be misplaced in the Xanthophyceae, and a selection of these taxa are the subject of this paper.
Most important for an improved definition of the Xanthophyceae was the realization that some of its traditional members constitute a group of their own, formalized as the class Eustigmatophyceae by Hibberd & Leedale (Citation1970, Citation1971). Over the years the number of taxa moved from xanthophytes to eustigmatophytes has been growing: more than half of the presently known ~35 eustigmatophyte species were originally placed in the Xanthophyceae (Hibberd, Citation1981; Eliáš et al., Citation2017; Amaral et al., Citation2020). Eustigmatophytes differ from xanthophytes by a suite of features concerning the ultrastructure and pigment composition, and are readily separated by molecular phylogenetics (Eliáš et al., Citation2017). However, the majority of traditional xanthophytes have not yet been studied using these modern approaches, and it is likely that a substantial proportion of them will prove to be hitherto unrecognized eustigmatophytes when investigated properly.
The genus Characiopsis Borzì is the largest genus formally classified in the Xanthophyceae (Ettl, Citation1978). It was established by Borzì (Citation1895) to accommodate species originally classified in the chlorophyte genus Characium Braun in Kützing (Citation1849), but that differed from bona fide members of the genus by accumulating oil as the reserve material rather than intraplastidial starch. Additional characters used by Borzì for distinguishing Characiopsis from Characium included the smaller number of chloroplasts and the absence of a pyrenoid. Six species were transferred by Borzì from Characium to Characiopsis, with Characium minutum Braun in Kützing, as Characiopsis minuta (Braun) Borzì, designated as the type species. Subsequent work by Lemmermann (Citation1914), Pascher (Citation1925, Citation1938), Ettl (Citation1960, Citation1977), Pizzaro (Citation1995), and others have substantially expanded the genus Characiopsis, partly by transfer from Characium, but mostly by describing new species. Generations of phycologists have generated some 190 names of Characiopsis species or their forms and varieties (Index Nominarum Algarum; https://ucjeps.berkeley.edu/ina). AlgaeBase, a key resource of taxonomic information for algae (Guiry & Guiry, Citation2020), presently lists 89 Characiopsis species flagged as accepted, together with 16 other species names of uncertain status or considered to be synonyms (https://www.algaebase.org/search/genus/detail/?genus_id=43814).
Interestingly, none of the Characiopsis species studied by modern methods has so far been confirmed as a xanthophyte; instead, they have all been demonstrated to belong to Eustigmatophyceae. The first such case was initially investigated under the name Ellipsoidion acuminatum and shown to exhibit the typical cytological features of eustigmatophytes (Hibberd & Leedale, Citation1970, Citation1972). The alga was later reidentified by Hibberd (Citation1981) as Characiopsis ovalis (Chodat) Chodat and thereafter treated as the new combination Pseudocharaciopsis ovalis (Chodat) Hibberd in the eustigmatophyte genus Pseudocharaciopsis Lee & Bold. Hibberd (Citation1981) additionally proposed that Pseudocharaciopsis texensis Lee & Bold, demonstrated to be a eustigmatophyte on the basis of its ultrastructure (Lee & Bold, Citation1973), is in fact synonymous with C. minuta, leading him to create a new combination Pseudocharaciopsis minuta (Braun) Hibberd. Molecular data confirmed that both Pseudocharaciopsis species are eustigmatophytes, but revealed that they are not directly related to each other, rendering the genus polyphyletic (Fawley et al., Citation2014). The 18S rRNA gene was subsequently sequenced from strains assigned to Characiopsis saccata N.Carter, Characiopsis acuta (Braun) Borzì, and Characiopsis longipes (Braun) Borzì (Fawley et al., Citation2014; Kryvenda et al., Citation2018), which proved to be eustigmatophytes closely related to P. minuta, and the placement of one of these strains in eustigmatophytes was further confirmed by a phylogenetic analysis of its plastid genome sequence (Ševčíková et al., Citation2019). However, no morphological data were provided for these strains and their identification was not verified, making the taxonomic implications of these findings uncertain.
Using molecular phylogenetics and transmission electron microscopy (TEM) we have recently studied several Characiopsis-like isolates, including strains identified as Characiopsis minima Pascher and Characiopsis aquilonaris Skuja (Amaral et al., Citation2020). Again, they were found to be eustigmatophytes, falling into a broader clade together with P. ovalis and another alga, Pseudellipsoidion edaphicum Neustupa & Němcová (Citation2001). This clade, previously known as the Pseudellipsoidion group (Fawley et al., Citation2014), was formalized as the new family Neomonodaceae, with a new genus Neomonodus created for (Pseudo)characiopsis ovalis. In addition, C. minima was transferred into a new genus, Characiopsiella, and C. aquilonaris was moved to the new genus Munda (Amaral et al., Citation2020). This work improved the classification of Characiopsis-like algae, addressing the polyphyly of the genus Pseudocharaciopsis, and by finding a home for two species from the apparently polyphyletic genus Characiopsis. Nevertheless, some key questions remain open: what is the actual position of the genus Characiopsis, and what is its relationship to the genus Pseudocharaciopsis?
To pursue the answer we present morphological and molecular characterization of 20 Characiopsis strains which have either not been studied before, or for which only molecular data have been reported. We combine the new findings with a discussion of the formal taxonomy of Characiopsis to conclude that this genus belongs to eustigmatophytes rather than xanthophytes and that Pseudocharaciopsis is a junior synonym of Characiopsis.
Materials and methods
Algal cultures and light microscopy
All strains selected for the study () were obtained from the Coimbra Collection of Algae (ACOI). The strains had previously been identified at ACOI based on light microscopy observations and attributed to the genus Characiopsis according to Ettl (Citation1978), Pizzaro (Citation1995), and original sources (when accessible to us) for species not covered by these two monographs. In addition, the strain ACOI 307 (Chlorobotrys regularis) was used to obtain the rbcL gene sequence (GenBank accession number MT374821) in order to improve the sampling for a phylogenetic analysis. The strains were cultivated in liquid Desmideacean Medium (Schlösser, Citation1994), pH 6.4–6.6, at 20°C, under a light intensity of 10 µmol photons m–2 s–1 (12:12 h photoperiod) provided by cool white fluorescent lamps. Morphological evaluation of the cells was performed using a Leica DMRB microscope with conventional light microscopy or DIC microscopy, using 60× and 100× PLAN APO objectives. Micrographs were acquired with a Nikon DS-Fi2 digital camera. Cell size was accessed by using the digital image analysis software NIS 4.60 (Isaza).
Table 1. Characiopsis strains studied and their characteristics. Cell morphology refers to the cell forms that predominate in the strains studied, with the caveat that the morphology can vary in the same culture. Dimensions of the cell body and stipe correspond to length × width (the general range is indicated, with exceptional values in parentheses)
Transmission electron microscopy
For TEM, the cell suspension was fixed for 150 min with 2.5% glutaraldehyde in phosphate buffer (0.05 M, pH 6.8), with glutaraldehyde subsequently washed out with the same buffer by centrifugation for 5 min at 2000 rpm. The cell suspension was embedded in 2% agar and post-fixed in 1% osmium tetroxide solution (prepared 1:1 (v/v) with the same phosphate buffer) for 2 h. The fixative was then washed out three times successively by addition of the buffer and centrifugation (5 min at 2000 rpm). Samples were dehydrated in an ethanol series (70%, 80%, 95% and 100%), each for 15 min. Samples were then embedded into a sequence of a mixture of ethanol and Spurr’s resin (33%, 50% and 66%) for 1 h and finally into 100% Spurr’s resin and kept overnight in a desiccator. Resin blocks were cut with an ultramicrotome (Ultracut E, Reichert-Jung) and ultrathin sections were mounted on copper grids and stained with 2% uranyl acetate and 0.2% lead citrate. Samples were examined in an FEI-Tecnai G2 Spirit Bio Twin electron microscope. Direct preparations of zoospores were obtained by fixing a drop of zoospore suspension on a formvar/carbon-coated grid in 2% osmium tetroxide vapour, drying at room temperature, and shadow casting with gold/palladium.
PCR amplification and DNA sequencing
Cells were collected by centrifugation of 2 ml of culture at 14 000 rpm and disrupted using a mixer mill (MM200, Retsch, Haan, Germany) and glass beads for 5 min. Genomic DNA was extracted using an Invisorb® Spin Plant Mini Kit (Stratek). PCR was performed with MyTaq™ Red DNA Polymerase (Bioline, UK) or Supreme NZYTaq II 2x Green Master Mix (Nzytech, Portugal), under the following conditions: denaturation at 95°C for 2 min, 35 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 2.5 min, and final extension at 72°C for 5 min. PCR products from amplification of the 18S rRNA and rbcL genes were purified using a GenElute™ PCR Clean-Up Kit (SIGMA). Sequences were obtained using a BigDye® Terminator v3.1 Cycle Sequencing Kit (ThermoFisher scientific) and analysed using the 3130xl Genetic Analyzer in the DNA Sequencing Laboratory of the Faculty of Science, Charles University (Prague). Primers used for obtaining full sequences of the 18S rRNA gene included the amplification primers 18S-F and 18S-R, and internal sequencing primers according to Katana et al. (Citation2001). Primers used for amplification of rbcL were EU-rbcL-F1 or eustigrbcL-F and the reverse primer EU-rbcL-R1 (Amaral et al., Citation2020), or alternatively a combination of the forward DPrbcL7 (Jones et al., Citation2005) and reverse NDrbcL8 (Daugbjerg & Andersen, Citation1997). For sequencing reactions, the amplification primers were used along with the newly designed sequencing primers (Amaral et al., Citation2020). Sequencing reads were assembled with SeqAssem (SequentiX, http://www.sequentix.de/software_seqassem.php) and manually edited by visual inspection of sequencing chromatograms. Sequences were trimmed to exclude primer regions and deposited at GenBank (accession numbers provided in ).
Phylogenetic analyses
The complete dataset for analyses of the 18S rRNA gene sequences included a total of 129 sequences and consisted of the 18 newly obtained sequences of Characiopsis strains, and a selection of 15 sequences from phylogenetically diverse ochrophytes to provide an outgroup. The sequences were aligned with MAFFT 7.429 (Katoh & Frith, Citation2012; Katoh & Standley, Citation2013), using the ‘Add’ option and a pre-existing alignment used in a previous study (Amaral et al., Citation2020). Redundant sequences were removed in BioEdit version 7.0.5 (Hall, Citation1999; http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and the resulting final alignment was trimmed with trimAl v1.4.rev6 by removing columns with more than 20% gaps (Capella-Gutiérrez et al., Citation2009; https://www.genome.jp/tools/ete/), leaving 1759 positions for tree inference. For the rbcL gene analysis, a selection of 74 eustigmatophyte sequences available from GenBank (retaining one sequence per described species for non-Characiopsis representatives) and the 20 newly obtained sequences were aligned with MAFFT 7.429. The termini of the alignment were trimmed in BioEdit to remove positions with a high percentage of missing data, leaving 1347 positions. Trees were inferred using the Maximum likelihood (ML) method implemented in RAxML (8.2.12) at the Cyberinfrastructure for Phylogenetic Research (CIPRESS) Portal (http://www.phylo.org/sub_sections/portal) (Miller et al., Citation2010) using the strategy of Stamatakis et al. (Citation2008) for obtaining the highest likelihood tree. The evolutionary model used was the default GTR+Γ. Bootstrap analyses were performed with the rapid bootstrapping procedure, with the adequate number of replicates detected by the program itself (‘halt bootstrapping automatically’ option); the number of bootstraps replicated for each tree is specified in the respective figure legends. Trees were drawn with the aid of the iTOL tool (Letunic & Bork, Citation2016; https://itol.embl.de/).
Results
General morphology of Characiopsis spp. strains
Vegetative cells of investigated Characiopsis are light green, with cell shapes varying from ovoid, fusiform (acute), ellipsoidal to cylindrical (–). Cell size varies from small cells 12–29 × 3–14 μm (rarely up to 63 μm long) to larger species 32–43 × 10–12 μm (rarely up to 84 μm long); old cells are wider and rounded up. All strains exhibit cell polarity with an attaching stipe or disc (stipitate cells) positioned at the posterior end of the cell. The stipe is an extension of the cell wall and is usually short (), but in some cases long and thin (). An orange-brownish accumulation occasionally observed on the stalk () possibly corresponds to mineral deposits containing manganese and other elements, as reported for Pseudocharaciopsis minuta (Wujek, Citation2012). On the apical end, cells may be round () or acute (, ) and often display a translucent tip (, ). The Characiopsis cell wall is smooth and continuous with the stalk and the apical tip (). Sometimes there was a visible thickening of the cell wall at the apical end and/or at the base () and it may look more like a distinctive refringent portion of the cell wall (), a feature also mentioned for Characiopsis naegelii Braun by Carter (Citation1919).
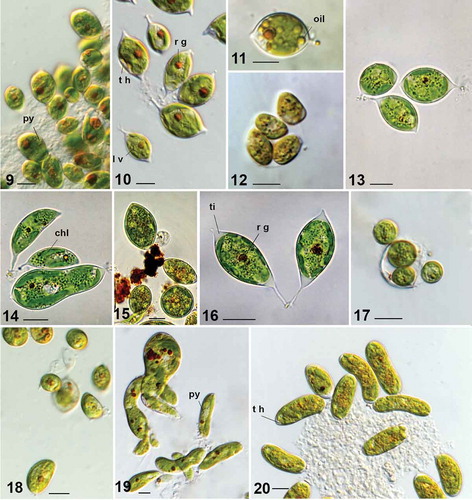
Figs 1–8. Characiopsis strains with morphology similar to Characiopsis minuta and unidentified Characiopsis strains. Fig. 1. Characium minutum (adapted from Braun, Citation1855). Fig. 2. Characiopsis minuta ACOI 2423. Fig. 3. Characiopsis cf. minuta ACOI 2425. Fig. 4. Characiopsis sp. ACOI 2432. Fig. 5. Characiopsis sp. ACOI 2438B. Fig. 6. Characiopsis sp. ACOI 2429. Fig. 7. Characiopsis sp. ACOI 2429A. Fig. 8. Characiopsis sp. ACOI 2430. Old cell (o c), pyrenoid (py), reddish globule (r g), stipe (st), zoospore (z), zoosporangium (zs). Photos 2–10 DIC 100× APO. Scale bars: 10 μm

Figs 9–20. Characiopsis strains with morphology unlike that of Characiopsis minuta. Fig. 9. Characiopsis pernana ACOI 2433. Figs 10–12. Characiopsis acuta ACOI 456. Fig. 13. Characiopsis acuta ACOI 1837. Fig. 14. Characiopsis longipes ACOI 1838. Fig. 15. Characiopsis longipes ACOI 1838. Fig. 16. Characiopsis longipes ACOI 2438. Fig. 17. Characiopsis longipes ACOI 1839_9. Fig. 18. Characiopsis cf. minutissima ACOI 2427A. Fig. 19. Characiopsis cf. saccata ACOI 481. Fig. 20. Characiopsis cedercreutzii ACOI 3169. Apical tip (ti), chloroplast (chl), lamellate vesicles (l v), oil droplets (oil), pyrenoid (py), reddish globule (r g), cell wall thickening (t h). Photos 11–14 and 19–22 DIC 100× APO. Scale bars: 10 μm
There are one to two parietal chloroplasts in young cells () and several in older cells. A globular, bulging pyrenoid was observed with light microscopy (, and ), as a refractile body projecting from the chloroplast. This structure was clearly seen in all investigated strains except for ACOI 2438, ACOI 3169 and ACOI 2436. A narrow stalk attaching the pyrenoid to the chloroplast was observed in sections (), in accordance with previous reports for P. minuta (Lee & Bold, Citation1973; Santos, Citation1996). The pyrenoid matrix is devoid of thylakoids and is surrounded by the characteristic flattened lamellate vesicles (). What appears to be a multiple-stalked pyrenoid may be seen in some sections ().
Figs 21–24. TEM sections of Characiopsis vegetative cells. Fig. 21. C. minuta ACOI 2423. Figs 22–24. C. cf. saccata ACOI 481. Chloroplast (chl), Golgi body (G b), lamellate vesicles (l v), mitochondrion (m), nucleus (n), nucleolus (nu), pyrenoid (py), reddish globule (r g). Scale bars: 1 μm
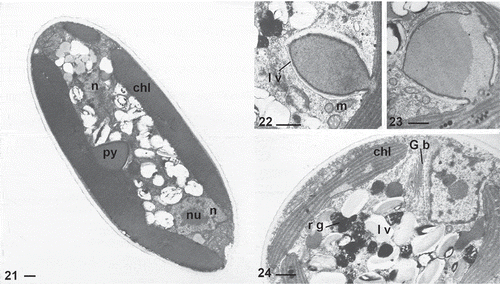
The two most distinctive eustigmatophyte organelles were found in the cells, the reddish globule (, , ) composed of several adjacent droplets not bound by any membrane and refractive lamellate vesicles scattered in the cytoplasm (). Other eukaryotic cell organelles were detected in TEM sections, such as a Golgi body lying next to the nucleus () and mitochondria with tubular cristae (). In larger cells more than one nucleus was present (), agreeing with previous studies of Munda sp. ACOI 2424 (Amaral et al., Citation2020), Neomonodus ovalis (syn. Characiopsis ovalis) (Poulton, Citation1926) and Characiopsis saccata (Carter, Citation1919). A connection between the chloroplast endoplasmic reticulum and the nuclear envelope could not be seen with confidence in any strain investigated by TEM and may be absent as generally seems to be the case in eustigmatophytes (; Eliáš et al. Citation2017). Reserves in the form of oil droplets were occasionally observed ().
As is generally true in eustigmatophytes (Eliáš et al. Citation2017; Amaral et al., Citation2020), only asexual reproduction was observed in Characiopsis spp. Abundant zoospore production (more than 10 per zoosporangium) was observed in the strains ACOI 2432 () and ACOI 2430. Despite their small size, zoospores were occasionally observed by light microscopy swimming with a long flagellum. Formation and release of four autospores per mother cell was observed ( and ).
Identification and specific characteristics of Characiopsis spp. strains
The original identification of the investigated strains at ACOI was re-evaluated, leading to revised identification in some cases. Ten strains could be matched with reasonable confidence to known Characiopsis species, whereas the other 10 strains displayed characteristics precluding their unambiguous identification at the species level. Identification and main morphological characteristics of the strains studied by us are summarized in .
Several strains were initially considered as candidates for an alga matching the original verbal description of Characium minutum by Braun (in Kützing, Citation1849), later documented in a drawing (Braun, Citation1855, see fig. 1). Of these, the strain ACOI 2423 seems to best fit the characteristics of the alga observed by Braun, and is thus identified here as Characiopsis minuta. The cells are long, acute, or frequently with a tip, and with a short stipe at the opposite end. They may be slightly curved on one or both ends, and become wider and larger in older cultures (). The cell size ranges from 12–29 (less frequently up to 42) µm in length and from 3 to 14 µm in width. The strain ACOI 2425 () is generally similar to ACOI 2423, but cells are longer and thinner than ACOI 2423. Owing to these differences, we cautiously refer to it as Characiopsis cf. minuta.
The strain ACOI 2432 would fit the morphological characteristics of C. minuta, but is noticeably larger (). The formation of more than 10 small zoospores per zoosporangium was observed (). Cells are larger than those of C. minuta, and most have a round rather than pointed apex (). The strains ACOI 2429 (), ACOI 2429A () and ACOI 2430 () also somewhat resemble C. minuta in morphology, but are wider and have a more rounded cell end (). Unambiguous identification to the species level based on all sources available to us proved impossible for strains ACOI 2432, 2438B, 2429, 2429A and 2430, so they are all referred to as Characiopsis sp.
Strain ACOI 2433 exhibits oval vegetative cells with a round apex and a short stipe (). It seems to be identifiable as Characiopsis pernana Pascher. Cells of ACOI 2427 and ACOI 2427A are small and wide, with a rounded apex, or sometimes acute and with a short stipe on the opposite end (). They resemble Characiopsis minutissima, but are significantly larger: 13–24(56) × 4–7(18) µm and 13–18(31) × 4–6(14) µm respectively, compared with the 6–9 × 4–6 µm reported for C. minutissima (Ettl, Citation1978). Therefore, we refer to them as Characiopsis cf. minutissima. One group of six strains is characterized by the presence of a long thin stipe. Two of them, ACOI 456 (–) and ACOI 1837 (), have oval cells with a tip. These characteristics fit the description of Characiopsis acuta. The strains ACOI 1838, 1839, 1839A and 2438 have longer oval (oblong) acute cells, with or without a tip, and the stipe is particularly long (6–12 µm) (, ; ACOI 1839 and 1839A are not shown). These ‘typical’ cells indicate that the strains correspond to the species Characiopsis longipes, although we note that other cell morphologies were observed to co-exist in the cultures.
The 18S rRNA gene sequence from the strain ACOI 481 has been reported (Kryvenda et al., Citation2018) and its replica SAG 15.97 (Fawley et al., Citation2014) was assigned to Characiopsis saccata, but without providing any data on the morphology of the strains. The cells are large, much longer than they are wide, with a short stipe at the base and rounded at the opposite end (). Some cells are straight, but the majority exhibit curved ends and widening of one or both ends of the cell, or even a contorted shape. Rarely, cells with a triangular form were found, too. Many reddish globules were found in the cells (). The cells differ from C. saccata in the fact that most cells do not have an acute end and also contorted cells are frequently seen, whereas the C. saccata cells depicted by Carter (Citation1919) are acute and not contorted, although some are curved. Owing to these doubts, we refer ACOI 481 (SAG 15.97) to Characiopsis cf. saccata. In strains ACOI 2434 and ACOI 3169, the cells are cylindrical, long and large, with a rounded apex. A thickening of the cell wall was sometimes observed in ACOI 3169 (). We confirm the initial identification of these two strains as Characiopsis cedercreutzii. The strain ACOI 2436, isolated from the same field sample as ACOI 2434, has shorter and wider cells (), so we leave it unidentified at the species level.
Molecular phylogeny of the Characiopsis strains
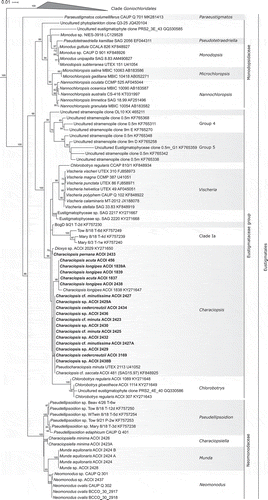
Altogether we report 18 new 18S rRNA gene sequences and 20 new rbcL gene sequences (). Together with the previously published data, both 18S rRNA and rbcL gene sequences are now available for 20 Characiopsis strains. The topology of the 18S rRNA tree () is congruent with the results of other recent analyses of this phylogenetic marker (Kryvenda et al., Citation2018; Ševčíková et al., Citation2019; Amaral et al., Citation2020). Two principal eustigmatophyte lineages are resolved in the tree, corresponding to the clade Goniochloridales (Fawley et al., Citation2014) and the order Eustigmatales. The latter is comprised of Paraeustigmatos columelliferus CAUP Q 701 (Fawley et al., Citation2019) branching off with moderate support as the most basal lineage, two strongly supported clades each corresponding to a formally recognized family, Monodopsidaceae (Hibberd, Citation1981) and Neomonodaceae (Amaral et al., Citation2020), and another strongly supported clade matching the previously defined Eustigmataceae group (Fawley et al., Citation2014). All 20 strains investigated in this study are placed in the Eustigmataceae group in a broader unsupported clade that also includes four previously sequenced Characiopsis strains and representatives of three additional nominal genera, Pseudocharaciopsis minuta UTEX 2113, Dioxys sp. ACOI 2029, and several Chlorobotrys isolates. Two subclades with strong or medium support emerge within this broader clade, one comprising all C. acuta and C. longipes strains and the other including most other strains except for C. pernana ACOI 2433 and Dioxys sp. ACOI 2029. The sequences of different strains within the two subclades are completely identical.
Fig. 25. Phylogeny of Eustigmatophyceae based on sequences of the 18S rRNA gene, showing the Eustigmatales. The phylogeny shown was inferred using the Maximum likelihood method implemented in RAxML (employing GTR+Γ substitution model) with bootstrap analysis followed by a thorough search for the ML tree. Bootstrap values higher than 50 are shown. Labels at terminal leaves comprise the strain updated taxonomic name followed by the collection reference number and the GenBank accession number. New sequences are highlighted in bold. The tree was rooted using 15 sequences from stramenopile algae sampled from GenBank. The outgroup is omitted and the ordinal clade Goniochloridales is shown collapsed for simplicity
The tree obtained using eustigmatophyte rbcL sequences () is likewise congruent with previous similar analyses (Ševčíková et al., Citation2019; Amaral et al., Citation2020), and divides eustigmatophytes into the same main lineages as the 18S rRNA gene tree. All of the newly investigated strains, together with the previously sequenced C. acuta ACOI 456, constitute a single, strongly supported clade (bootstrap value 99%) within the Eustigmataceae group, which we hereafter call the Characiopsis clade. More genetic variation is recorded in the rbcL gene compared with the 18S rRNA gene, allowing better resolved relationships both within the Eustigmataceae group and the Characiopsis clade. Thus, the clade Ia comprised of several unidentified isolates (Fawley et al., Citation2014) is positioned with strong support as a lineage sister to the other representatives of the Eustigmataceae group for which the rbcL gene sequence is available. Chlorobotrys regularis ACOI 307, newly sequenced by us to improve the sampling of the Eustigmataceae group, belongs to a moderately supported group together with the genus Vischeria (including the former Eustigmatos species; Kryvenda et al., Citation2018), whereas the Characiopsis clade may be specifically related to the unidentified isolate BogD 9/21 T-2d, although this relationship is supported only by a moderate bootstrap value (70%).
Fig. 26. Phylogeny of Eustigmatophyceae based on the rbcL gene, showing the Eustigmatales. The phylogeny shown was inferred using the Maximum likelihood method implemented in RAxML (employing GTR+Γ substitution model) with bootstrap analysis followed by a thorough search for the ML tree. Bootstrap values higher than 50 are shown. Labels at terminal leaves comprise the strain updated taxonomic name followed by the collection reference number when applicable and the GenBank accession number. New sequences highlighted in bold. The tree was rooted at the ordinal clade Goniochloridales which is shown collapsed for simplicity
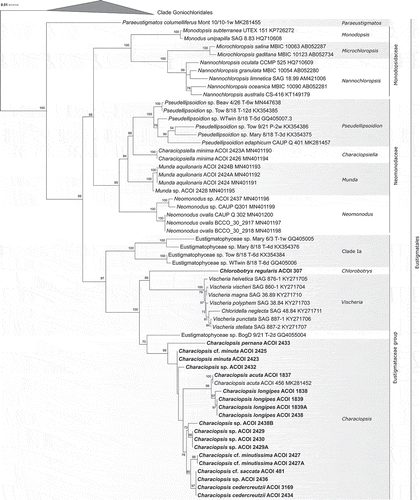
The internal structure of the Characiopsis clade in the rbcL tree is more elaborate than the corresponding part of the 18S rRNA tree, although many deep branches lack statistical support (). Strains of C. acuta and C. longipes constitute a clearly delimited strongly supported (99% bootstrap) clade separated from other Characiopsis strains by a long stem branch. Some sequence heterogeneity is apparent among C. longipes strains, with ACOI 1838 being separated from the other three strains. Another noticeable grouping comprises ACOI 481 (Characiopsis cf. saccata), ACOI 2436 (Characiopsis sp.) and ACOI 2434 and 3169 (C. cedercreutzii), with identical rbcL sequences. This cluster is related (80% bootstrap support) to the two strains referred to as C. cf. minutissima (ACOI 2427 and 2427A), and together with them may belong to an even more inclusive group (78% bootstrap support), additionally embracing four unidentified Characiopsis strains. Three of these strains share identical rbcL sequences and are therefore likely to be conspecific. The rbcL sequences of ACOI 2423 (C. minuta) and ACOI 2425 (C. cf. minuta) are also identical.
Discussion
Identity of the genus Characiopsis
In this paper we provide morphological and molecular data for a series of algal strains identified as species of Characiopsis, or at least fitting the general characteristic morphology of this genus. All the strains proved to be eustigmatophytes, specifically members of the Eustigmataceae group. This position is also consistent with the phylogenetic analysis of concatenated plastid genome-encoded proteins including a single representative of the Characiopsis clade, C. acuta ACOI 456 (Ševčíková et al., Citation2019). Whereas the eustigmatophyte nature of these algae is undeniable, the question arises of whether this also implies that the genus Characiopsis as such should be transferred from its current taxonomic home, the class Xanthophyceae, to the class Eustigmatophyceae.
The answer relies on resolving the actual identity of the type of the genus. However, what is to be considered the type of Characiopsis Borzì has become a matter of controversy in the literature. When establishing the genus, Borzì (Citation1895, p. 154) explicitly stated that ‘Tuttavia una forma sulla quale non parmi possano cadere di dubbi e che con contezza debba assumersi come tipo del nuovo genere Characiopsis è il Characium minutum di Al. Braun’. In the same publication Borzì provided drawings of an alga he observed and identified as C. minutum Braun, designated by him with the new combination Characiopsis minuta. However, Lemmermann (Citation1914) had an opportunity to study the original specimen used by Braun to describe C. minutum, and based on this he concluded that the alga documented by Borzì is a different species, which he described as Characiopsis borziana Lemmermann. In light of this, Silva (Citation1979) interpreted the typification of Characiopsis as follows (p. 40): ‘In my opinion a genus should be typified with material at hand, whether or not the author misidentified the type with a previously described species. Accordingly, I consider C. borziana the type of its genus’. This interpretation was adopted by Hibberd (Citation1981), who considered it in agreement with the intention of Article 10.1 of the International Code of Botanical Nomenclature, and later also by Pizarro (Citation1995).
However, after years of debating by authorities on botanical nomenclature (McNeill, Citation1981), a modified version of Article 10.1 appeared in the 1983 edition of the Code and remains the same in the current edition of the International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) (Turland et al., Citation2018). The Article specifically states: ‘The type of a name of a genus or of any subdivision of a genus is the type of a name of a species…For purposes of designation or citation of a type, the species name alone suffices, i.e. it is considered as the full equivalent of its type’. Hence, according to the current meaning of the Code the type of the genus name Characiopsis is the type of the species name Characium minutum Braun, regardless of what exactly Borzì had at hand and identified as C. minutum. Braun’s description of the species is not accompanied by an illustration and does not explicitly specify a type (Kützing, Citation1849, p. 892). However, Lemmermann (Citation1914) mentioned Braun’s ‘Original exemplare von Ch. minuta’ in Berlin Herbarium that he could study (see above), which can be considered the holotype of C. minutum Braun. Unfortunately, this specimen no longer exists and was most likely destroyed during World War II (Dr Nélida Abarca, Botanic Garden and Botanical Museum Berlin, personal communication). Hence, following Article 9.16 of the Code, we here designate a neotype for C. minutum Braun. Specifically, we select Braun’s original drawing of a particular cell of C. minutum published by him in 1855 and reprinted here as part of (no. 6). It then follows that this drawing represents the type of the genus name Characiopsis.
Characiopsis minuta is a eustigmatophyte
The key step towards final resolution of the question of which lineage should be called Characiopsis and where it fits phylogenetically is to investigate an alga that can unambiguously be identified as Braun’s C. minutum. Several strains from our set were considered as possible candidates, including those initially identified by the ACOI curator as C. minuta (ACOI 2423 and ACOI 2425) and ‘Characiopsis minuta?’ (ACOI 2429 and 2429A). A careful re-evaluation of the morphology of these strains led us to conclude that ACOI 2423 best fits the characteristics of C. minuta and can be used as a basis for further taxonomic reasoning concerning the genus Characiopsis.
Another strain was previously proposed to represent C. minuta. Hibberd (Citation1981) discussed in detail the morphology of the authentic strain of the eustigmatophyte alga Pseudocharaciopsis texensis Lee & Bold and concluded that it can be identified as conspecific with Characiopsis minuta. Given his conviction that Characiopsis is typified by C. borziana (see above) and that the genus should stay in Xanthophyceae, he created a new combination Pseudocharaciopsis minuta, with P. texensis as its junior synonym. The sequence of the 18S rRNA gene of this strain (held in the UTEX collection as the culture UTEX 2113) was obtained by Andersen et al. (Citation1998) and confirms that this alga is indeed closely related to the studied Characiopsis strains, including the proposed candidate for C. minuta, the strain ACOI 2423 (, ). In fact the 18S rRNA sequences of these two strains differ by only two single nucleotide indels, and inspection of a multiple alignment of available eustigmatophyte sequences revealed that the differences map into conserved regions of the gene, with the UTEX 2113 sequence being the one that deviates from the conserved pattern (not shown). Considering that this sequence was obtained by manual sequencing on a polyacrylamide gel (Andersen et al., Citation1998), a less accurate procedure than the current Sanger method, the differences between this sequences and those obtained from other eustigmatophytes might possibly be artefacts. Unfortunately, the strain is no longer available from the UTEX collection, and an equivalent strain held in the CCAP collection with the reference number 864/1 (see Hibberd, Citation1981) is likewise lost (overgrown by a green alga; details not shown). It is not therefore possible to sequence its 18S rRNA gene once more, nor to determine the sequence of its rbcL gene to get a more precise understanding of its relationship to strains investigated in the present study. Nevertheless, the evidence available is compatible with a notion that the strain ACOI 2423 is closely related, if not conspecific, with what Hibberd interpreted as C. minuta. Consequently, our results strengthen the argument for the name P. texensis being considered synonymous with C. minuta.
To stabilize the meaning of the name Characiopsis minuta, and thus to anchor the definition of the genus Characiopsis, below we designate an epitype to support the neotype of Characium minutum (Braun’s drawing, see above), whose practical use as a reference for identification of the species is inherently limited. Strain ACOI 2423 is morphologically the closest among available Characiopsis strains to the alga reported by Braun as Characium minutum, and we have confidence in its identification as the same species. Hence, a metabolically stable cryopreserved material derived from a living culture of this strain is here designated as the epitype of Characium minutum (= Characiopsis minuta).
Characiopsis is a eustigmatophyte genus; Pseudocharaciopsis is its junior synonym
Based on the above data and arguments, the question of the identity of the type of the genus Characiopsis is resolved. Also, with the convincingly demonstrated position of the type in a particular lineage of eustigmatophytes, the debate on whether the genus should be classified in Xanthophyceae or Eustigmatophyceae seems to be closed. However, Hibberd (Citation1981) discussed the morphological features of the type species of the genus Characiopsis, C. borziana, and considered the possibility that, like C. minuta, it may also be a eustigmatophyte, even possibly congeneric with Pseudocharaciopsis species. Should this prove to be the case, he proposed that the ‘transfer of the name Characiopsis from the Tribophyceae to the Eustigmatophyceae could be prevented either by conserving Characiopsis with an altered type or by proposing the name as a nomen rejiciendum on the grounds that it had been widely and persistently used for a taxon not including its type’ (Hibberd, Citation1981, p. 109). So, when confronted with the situation de facto envisaged by Hibberd, should we consider implementing his formal taxonomic act to preserve Characiopsis as a genus of xanthophyte algae and Pseudocharaciopsis as an independent genus in eustigmatophytes?
One argument against this advice is that all representatives of the traditionally circumscribed genus Characiopsis studied so far by modern methods prove to be eustigmatophytes rather than xanthophytes. In addition to C. minuta, this was previously demonstrated for C. ovalis, C. minima and C. aquilonaris, which are however not directly related to the ‘main’ Characiopsis clade (see also , ) and each has been placed in its own newly erected genus (Neomonodus, Characiopsiella and Munda, respectively; Amaral et al., Citation2020). The present study adds four known morphologically recognized () and up to nine genetically delimited Characiopsis species (see below) that are eustigmatophytes related to C. minuta. Hence, at the moment there is no strong evidence for any nominal Characiopsis species being a xanthophyte. So, Hibberd’s proposal currently lacks support. With the present knowledge of Characiopsis-like algae we can conclude that Characiopsis is a genus embracing a set of closely related eustigmatophyte algae in the Eustigmataceae group, and that Pseudocharaciopsis is its junior synonym.
This conclusion does not necessarily imply that all algae presently classified in the genus Characiopsis must belong to Eustigmatophyceae. In this regard it is interesting to consider a note (Lee & Bold, Citation1973, p. 37) in their paper describing P. texensis: ‘The writers have in their collection two strains of Characiopsis. It was of interest to ascertain whether these also should be assigned to the new genus Pseudocharaciopsis. However, both light and electron microscopy indicated that they are not eustigmatophycean algae.’ Unfortunately, no further details on these strains seem to have been published by the authors and it is unclear whether they represented xanthophytes or yet another algal group. While the morphological characters documented for the various Characiopsis species are generally insufficient to determine their actual affiliation, some species seem unlikely to belong to Eustigmatophyceae and some may indeed be xanthophytes instead. For example, zoospores in Characiopsis elegans and Characiopsis galeata were depicted with a laterally positioned eyespot located in the chloroplast (Ettl, Citation1956), which contrasts with the characteristic zoospore structure in eustigmatophytes featuring an extraplastidial anterior eyespot associated with the base of the long flagellum (Hibberd, Citation1981). A renewed culturing effort will hopefully enable re-evaluation of a broader set of Characiopsis species with molecular and other modern methods of algal systematics, which may ultimately unveil Characiopsis-like species belonging to xanthophytes or other classes outside eustigmatophytes. Should such species be found, they will have to be transferred from Characiopsis to a new genus or genera.
Phylogenetic delimitation of the genus Characiopsis
Phylogenetic analyses of both 18S rRNA and rbcL gene sequences generally support the existence of a eustigmatophyte subgroup including C. minuta and a suite of genetically more or less differentiated strains, interpreted as the genus Characiopsis (, ). The rbcL tree shows the genus as a strongly supported clade well separated from other eustigmatophyte lineages. In contrast, the 18S rRNA gene tree does not exhibit an equivalent clade due to a cluster of Chlorobotrys sp. sequences nested among Characiopsis sp. sequences. An analogous grouping, denoted ‘Pseudocharaciopsis/Chlorobotrys/Dioxys-clade’, was also retrieved in a previous study with a much poorer sampling of Characiopsis diversity (Kryvenda et al., Citation2018). The position of the Chlorobotrys cluster in the 18S tree, not supported by the bootstrap analysis (), is incongruent with the position of a representative of this cluster (C. regularis ACOI 307) in the rbcL tree, where it is placed (with 86% BP) sister to the genus Vischeria (). Instead, the unidentified strain Bog 9/21 T-2d may be more closely related to Characiopsis according to rbcL data.
The 18S rRNA gene provides insufficient resolution for the branching order at the base of the Eustigmataceae group, and even for demonstrating the monophyly of Characiopsis. There is a noticeably lower rate of evolution of the 18S rRNA gene in Characiopsis spp. as compared with most other members of the Eustigmataceae group: note the short branches of the Characiopsis sequences and their identity or high similarity even in species that are well differentiated by morphology and rbcL sequences. As a result, probably only a few synapomorphic mutations have accumulated in the 18S rRNA gene of the Characiopsis lineage, allowing for robust inference of its monophyly. While the rbcL gene phylogeny and morphological characters collectively provide sufficient support for the delimitation of the genus Characiopsis as a monophyletic entity within the Eustigmataceae group, multigene analyses additionally including lineages currently represented only by 18S rRNA data (such as the lineage comprised of the two unidentified strains SAG 2217 and 2220) are required to better understand the phylogenetic position of Characiopsis among its closest relatives.
Interestingly, the 18S rRNA tree suggests that specific relatives of the nominal Characiopsis strains (including the authentic Pseudocharaciopsis texensis strain reinterpreted as C. minuta, see above) may also include the strain ACOI 2029 assigned as an unidentified species in the genus Dioxys (). The respective sequence was obtained by Kryvenda et al. (Citation2018), but the authors did not provide information on the morphology of Dioxys sp. ACOI 2029, so the validity of the identification remains uncertain. However, the genus Dioxys Pascher exhibits clear resemblance to Characiopsis owing to the presence of a stipe, and the probable specific relationship of the two genera is reflected by their classification into the informal ‘Characiopsis-Gruppe’ in the family Characiopsidaceae (Ettl, Citation1978). It cannot be ruled out that some Dioxys species in fact belong phylogenetically to the Characiopsis clade and should be reclassified accordingly. If this also applied to the type species, Dioxys incus Pascher, Dioxys (described in Pascher, Citation1932) would have to be reconsidered as a junior synonym of Characiopsis. Future investigations of the Dioxys sp. ACOI 2029 strain will help test these possibilities.
Interestingly, the Characiopsis-like morphology is not restricted to a single evolutionary lineage of eustigmatophytes, as several species historically classified as Characiopsis are found in a group distantly related to the Characiopsis clade that was recently defined as the new family Neomonodaceae. These species, now placed in Neomonodus, Characiopsiella and Munda (, ), were noted to share a morphological feature discriminating them from Characiopsis and Pseudocharaciopsis species in the Eustigmataceae group: the absence of a pyrenoid (Amaral et al., Citation2020). Indeed, we observed a pyrenoid by light microscopy in nearly all studied (bona fide) Characiopsis strains (), further strengthening the case that the presence or absence of a pyrenoid is a phylogenetically informative character in algae with Characiopsis-like morphology. TEM sections may clarify if this structure is present in the three Characiopsis strains in which a pyrenoid could not be discerned under light microscopy. If confirmed, the absence of a pyrenoid would be a recently evolved feature of these strains, as they are all closely related (most likely conspecific) with strains that have it (, ).
Diversity in the genus Characiopsis
Whereas we have shown that the 18S rRNA gene evolves slowly in Characiopsis and hence is not particularly informative about the (phylo)genetic diversification within the genus (), rbcL gene sequences demonstrate considerable diversity (). A natural question is how this diversity translates into formal classification, i.e. delimitation of species within the genus. Considering the degree of differences in rbcL sequences between different nominal species in other eustigmatophyte genera, up to 10 separate species seem to have been captured by the present sampling of the genus Characiopsis (). Some interesting conclusions can be drawn when the molecular data are combined with morphological observations of the strains.
First, morphologically similar strains may prove to be genetically different, as is the case with the set of strains more or less reminiscent of C. minuta, and the strains identified as C. longipes. Thus, it is possible that new Characiopsis species need to be recognized to properly reflect the actual diversity within the genus. Second, the Characiopsis cultures usually exhibit a range of different cell morphologies, also depending on the age of the culture. This makes it difficult to match the organisms to the species descriptions provided by previous authorities, which were typically derived from observing the algae in natural samples and thus could not really capture the actual morphological plasticity the species exhibits in reality. Finally, morphologically distinguishable strains may be genetically so close as to be possibly conspecific. Most notable is the case of a cluster of four strains with identical rbcL sequences (), two of which identified as C. cedercreutzii, one resembling C. saccata and the fourth without clear species assignment (, ). We really do not understand whether this reflects the insufficiency of even the rbcL gene to discriminate closely related yet distinct species, or whether we have encountered a case of considerable morphological plasticity whereby the same species may look different depending on minor genetic or epigenetic differences.
Indeed, the life cycles of eustigmatophyte algae including Characiopsis are yet to be clarified, and we cannot, for instance, exclude the possibility that some of the eustigmatophytes exhibit alternation of generations, i.e. vegetative phases occurring at two different ploidy levels and potentially differing in their morphology. Another factor potentially impacting the appearance of the algae in culture are biotic interactions with co-cultivated microorganisms (bacteria, fungi etc.; note that the strains studied are not in axenic cultures). In this regard it is interesting to note that some eustigmatophytes, including two strains of C. acuta studied in this paper (ACOI 456 and ACOI 1837), were recently shown to harbour endosymbiotic bacteria representing a new genus of the family Rickettsiaceae (Candidatus Phycorickettsia; Yurchenko et al., Citation2018). To what extent the presence of the endosymbiont in the algal host influences its morphology is presently unknown, but some effects would not be surprising. With these arguments in mind we refrain from herein proposing taxonomic changes such as description of new species or synonymization of existing species, since we feel our understanding of the biology and phylogenetic diversity of Characiopsis is presently insufficient to make such an effort substantiated and well founded in the data.
Renewed culturing efforts combined with modern ‘omics’ approaches will be instrumental in improving our knowledge of the genus Characiopsis. Indeed, a genome survey has recently been conducted for C. acuta ACOI 456, yielding a complete plastid genome sequence (Ševčíková et al., Citation2019) and much more, including genome data from its Phycorickettsia endosymbiont (Eliáš et al., unpublished results). Our present work establishes a useful framework for future exploration of the biological mysteries of this fascinating algal group.
Formal taxonomy
Characiopsis minuta (Braun) Borzì
BASIONYM: Characium minutum Braun in Kützing (Citation1849)
HETEROTYPIC SYNONYM: Pseudocharaciopsis minuta (Braun) Hibberd
NEOTYPE (designated here): The single cell illustrated as no. 6 in Tab V, F in Braun (Citation1855); see also in this paper.
EPYTYPE (designated here to support the neotype): strain ACOI 2423 permanently preserved in a metabolically inactive state (cryopreserved in liquid nitrogen), deposited at ACOI – Coimbra Collection of Algae, University of Coimbra.
Note: While we consider the illustration cited above a neotype, it is formally possible that it should instead be designated as a lectotype. The latter would become appropriate should it be demonstrated that Braun in reality prepared the illustration before the publication of the species description in 1849. Such a possibility cannot be ruled out, but since Braun (Citation1855, p. 46) mentions his observations of C. minutum in 1851 and 1854, i.e. after the first encounter of the species in 1848, it is likely the drawing published in 1855 postdates the description in 1849. Hence, neotypification rather than lectotypification seems to be the appropriate act with the evidence available at the moment.
Author contributions
R. Amaral: conceived the study, obtained and interpreted the morphological and ultrastructural data, obtained and analysed sequence data, prepared figures and tables, drafted and edited the manuscript; M. Eliáš: conceived the study, obtained and analysed sequence data, drafted and edited the manuscript; L.M.A. Santos: conceived the study, obtained and interpreted the morphological and ultrastructural data, and edited the manuscript; T. Ševčíková obtained and analysed sequence data and edited the manuscript.
Acknowledgements
We acknowledge researcher Fátima Santos, former curator of the ACOI collection, for isolating the strains studied in this paper and for her initial insight concerning their identification. We thank an anonymous reviewer and Prof. Karol Marhold (Charles University, Prague) for very useful recommendations concerning the issues of formal taxonomy and nomenclature discussed in the paper, Dr Nélida Abarca (Botanic Garden and Botanical Museum Berlin) for information concerning the status of Braun’s original specimen of Characium minutum, and John Cawley for correcting our English.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Amaral, R., Fawley, K.P., Němcová, Y., Ševčíková, T., Lukešová, A., Fawley, M.W., Santos, L.M.A. & Eliáš, M. (2020). Towards modern classification of eustigmatophytes: Neomonodaceae, fam. nov., with the description of three new genera. Journal of Phycology, 56: 630–648.
- Andersen R.A., Brett R.W., Potter D. & Sexton J. (1998). Phylogeny of the Eustigmatophyceae based upon 18S rDNA, with emphasis on Nannochloropsis. Protist, 149: 61–74.
- Borzì, A. (1895). Characiopsis. In Studi algologici: saggio di richerche sulla biologia delle alghe. Vol. II (Reber, A., editor), 121–378. Libreria Carlo Clausen, Palermo.
- Braun, A. (1855). Characium. In Algarum Unicellularium Genera Nova et Minus Cognita: Praemissis Observationibus de Algis Unicellularibus in Genere (Engelmann, A., editor), 29–47. Lipsiae.
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25: 1972–1973.
- Carter, N. (1919). On the cytology of two species of Characiopsis. New Phytologist, 18: 177–186.
- Darienko, T., Gustavs, L., Mudimu, O., Menendez, C.R., Schumann, R., Karsten, U., Friedl, T. & Proschold, T. (2010). Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta). European Journal of Phycology, 45: 79–95.
- Daugbjerg, N. & Andersen, R. A. (1997). A molecular phylogeny of the heterokont algae based on analyses of chloroplast‐encoded rbcL sequence data. Journal of Phycology, 33: 1031–1041.
- Eliáš, M., Amaral, R., Fawley, K.P., Fawley, M.W., Němcová, Y., Neustupa, J., Přibyl, P., Santos, L.M.A. & Ševčíková, T. (2017). Eustigmatophyta. In Handbook of the Protists (Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J. & Corliss, J.O., editors), 1–39. Springer International Publishing, Cham.
- Eliáš, M., Neustupa, J., Pažoutová, M. & Škaloud, P. (2013). A case of taxonomic inflation in coccoid algae: Ellipsoidion parvum and Neocystis vischeri are conspecific with Neocystis (=Nephrodiella) brevis (Chlorophyta, Trebouxiophyceae). Phytotaxa, 76: 15–27.
- Ettl, H. (1956). Ein Beitrag zur Systematik der Heterokonten. Botaniska Notiser, 109: 411–455.
- Ettl, H. (1960). Neue Vertreter der Gattung Characiopsis. Botaniska Notiser, 113: 265–272.
- Ettl, H. (1977). Taxonomische Bemerkungen zu den Xanthophyceen. Nova Hedwigia, 28: 555–568.
- Ettl, H. (1978). Xanthophyceae. In Süsswasserflora von Mitteleuropa, Bd. 3. 1. Teil (Ettl, H., Gerloff, H.J. & Heynig, H., editors), 341–381. Gustav Fischer Verlag, Stuttgart.
- Fawley, K.P., Eliáš, M. & Fawley, M.W. (2014). The diversity and phylogeny of the commercially important algal class Eustigmatophyceae, including the new clade Goniochloridales. Journal of Applied Phycology, 26: 1773–1782.
- Fawley, M.W., Němcová, Y. & Fawley, K.P. (2019). Phylogeny and characterization of Paraeustigmatos columelliferus, gen. et sp. nov., a member of the Eustigmatophyceae that may represent a basal group within the Eustigmatales. Fottea, 19: 107–114.
- Gärtner, G. & Schragl, A. (1988). Zur taxonomie von Coccomyxa brevis nov. comb. (Chlorophyceae, Chlorococcales), füher Nephrodiella brevis Vischer (Xanthophyceae, Mischococcales). Nova Hedwigia, 46: 511–517.
- Guiry, M.D. & Guiry, G.M. (2020). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org.
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95–98.
- Hibberd, D.J. (1981). Notes on the taxonomy and nomenclature of the algal classes Eustigmatophyceae and Tribophyceae (synonym Xanthophyceae). Journal of the Linnean Society of London Botany, 82: 93–119.
- Hibberd, D.J. & Leedale, G.F. (1970). Eustigmatophyceae – a new algal class with unique organization of the motile cell. Nature, 225: 758–760.
- Hibberd, D.J. & Leedale, G.F. (1971). A new algal class – the Eustigmatophyceae. Taxon, 20: 523–525.
- Hibberd, D.J. & Leedale, G.F. (1972). Observations on the cytology and ultrastructure of the new algal class, Eustigmatophyceae. Annals of Botany, 36: 49–71.
- Hibberd, D.J. & Norris, R.E. (1984). Cytology and ultrastructure of Chlorarachnion reptans (Chlorarachniophyta divisio nova, Chlorarachniophyceae classis nova). Journal of Phycology, 20: 310–330.
- Jones, H.M., Simpson, G.E., Stickle, A.J. & Mann, D.G. (2005). Life history and systematics of Petroneis (Bacillariophyta) with special reference to British waters. European Journal of Phycology, 40: 61–87.
- Katana, A., Kwiatowski, J., Spalik, K., Zakryš, B., Szalacha, E. & Szymańdka, H. (2001). Phylogenetic position of Koliella (chlorophyte) as inferred from nuclear and chloroplast small subunit rDNA. Journal of Phycology, 37: 443–451.
- Katoh, K. & Frith, M.C. (2012). Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics, 28: 3144–3146.
- Katoh, K. & Standley, D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30: 772–780.
- Kryvenda, A., Rybalka, N., Wolf, M. & Friedl, T. (2018). Species distinctions among closely related strains of Eustigmatophyceae (Stramenopiles) emphasizing ITS2 sequence-structure data: Eustigmatos and Vischeria. European Journal of Phycology, 53: 471–491.
- Kützing, F.T. (1849). Species Algarum. F.A. Brockhaus (editor), 1–922. Leipzig.
- Lee, K.W.L. & Bold, H.C. (1973). Pseudocharaciopsis texensis gen. et. sp. nov., a new member of the Eustigmatophyceae. British Phycological Journal, 8: 31–37.
- Lemmermann, E. (1914). Algologische Beitrage. XII Die Gattung Characiopsis Borzì. Abhandlungen herausgegeben vom Naturwissenschaftlichen Verein zu Bremen, 23: 249–267.
- Letunic, I. & Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research, 44: W242–W245.
- Maistro, S., Broady, P., Andreoli, C. & Negrisolo, E. (2017). Xanthophyceae. In Handbook of the Protists (Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J. & Corliss, J.O., editors), 407–434. Springer, Cham.
- McNeill, J. (1981). Report of the committee on generic typification. Taxon, 30: 200–207.
- Miller, M.A., Pfeiffer, W.T. & Schwartz, T. (2010). Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Conference paper. In Proceedings of Gateway Computing Environments Workshop (GCE), New Orleans, Louisiana, USA. doi: 10.1109/GCE.2010.5676129.
- Neustupa, J. & Němcová, Y. (2001). Morphological and taxonomic study for three terrestrial eustigmatophycean species. Nova Hedwigia, 123: 373–386.
- Pascher, A. (1925). Heterokontae. In Die Süsswasser-Flora Deutschlands, Österreichs und der Schweiz, Bd. 11: Heterokontae, Phaeophyta, Rhodophyta, Charophyta. (Pascher, A., Schiller, J. & Migula, W., editors), 1–118. Verlag von Gustav Fischer, Jena.
- Pascher, A. (1932). Über einige neue oder kritische Heterokonten. (Beiträge zur Kenntnis der einheimischen Algenflora. II.). Archiv für Protistenkunde, 77: 305–359.
- Pascher, A. (1938). Heterokonten. In Kryptogamen-Flora von Deutschland, Österreich und der Schweiz (Rabenhorst, L., editor) Vol. 11, Teil 5, 641–832. Akademische Verlagsgesellschaft, Leipzig.
- Pizarro, H. (1995). The genus Characiopsis Borzi (Mischococcales, Tribophyceae). Taxonomy, biogeography and ecology. Bibliotheca Phycologica, 98: 1–113.
- Poulton E. (1926). Studies on the Heterokontae. New Phytologist, 25: 313–319.
- Santos, L.M.A. (1996). The Eustigmatophyceae: Actual knowledge and research perspectives. Nova Hedwigia, 112: 391–405.
- Schlösser, U.G. (1994). SAG- Sammlung von Algenkulturenat the University of Göttingen. Catalogue of strains. Botanica Acta, 107: 175–176.
- Ševčíková, T., Yurchenko, T., Fawley, K.P., Amaral, R., Strnad, H., Santos, L.M.A., Fawley, M.W. & Eliáš, M. (2019). Plastid Genomes and Proteins Illuminate the Evolution of Eustigmatophyte Algae and Their Bacterial Endosymbionts. Genome Biology and Evolution, 11: 362–379.
- Silva, P.C. (1979). Review of the taxonomic history and nomenclature of the yellow-green algae. Archiv fur Protistenkunde, 121: 20–63.
- Stamatakis, A., Hoover, P. & Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology, 57: 758–771.
- Turland, N.J., Wiersema, J.H., Barrie, F.R., Greuter, W., Hawksworth, D.L., Herendeen, P.S., Knapp, S., Kusber, W.-H., Li, D.-Z., Marhold, K., May, T.W., McNeill, J., Monro, A.M., Prado, J., Price, M.J. & Smith, G.F. (eds) (2018). Regnum Vegetabile. In International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017, 159. Koeltz Botanical Books, Glashütten.
- Wujek, D.E. (2012). Biomineralization on the stalk of the eustigmatophyte Pseudocharaciopsis (Eustigmatophyceae). Algae, 27: 135–137.
- Yurchenko, T., Ševčíková, T., Přibyl, P., El Karkouri, K., Klimeš, V., Amaral, R., Zbránková, V., Kim, E., Raoult, D., Santos, L.M.A. & Eliáš, M. (2018). A gene transfer event suggests a long-term partnership between eustigmatophyte algae and a novel lineage of endosymbiotic bacteria. ISME Journal, 12: 2163–2175.
