ABSTRACT
Many dinoflagellate groups have been overlooked because of their small size compared with their larger counterparts; consequently, little is known about their diversity, distribution, and contribution to the planktonic community. Ansanella is a recently described genus of small marine planktonic dinoflagellates belonging to the order Suessiales. In this study, morphological observations together with molecular SSU and LSU rDNA analyses of clonal strains from the Catalan coast (NW Mediterranean Sea) allowed the identification of A. cf. natalensis and a newly reported species, Ansanella catalana. The cells of A. catalana are oval and 9.6–15.5 μm long; the cell surface is covered with amphiesmal vesicles arranged in 10–13 latitudinal rows; the episome has an apical furrow complex with a single elongated apical vesicle; a type E eyespot is also present. This newly identified species can be distinguished from congeneric species by morphological details of the cell surface, the presence of mucocysts and an extrusome type not previously reported in dinoflagellates. The characterization of this structure could provide insights into extrusome evolution in dinoflagellates. Metabarcoding analysis of plankton and sediment samples from the Catalan coast revealed a wide distribution of Ansanella and the remarkable contribution of this genus to the dinoflagellate communities from three different coastal habitats.
Highlights
Members of Ansanella genus are widespread along the Catalan coast.
A new species of the genus Ansanella (Dinophyceae) is described.
A new type of extrusome was observed.
Introduction
Dinoflagellates are a diverse group of protists differing from one another in several of their morphological features, such as their tabulation and the types of their internal organelles, including extrusomes and eyespots. Although ~2500 species of dinoflagellates have been described to date (Hoppenrath et al., Citation2009; Gómez, Citation2012), the true species diversity is difficult to estimate (Hoppenrath, Citation2017). Consequently, the full spectrum of their morphological features, biological traits, life strategies and genetic variability remains unknown. Moreover, dinoflagellates are still largely identified based on light microscopy observations, but this method is not sufficient to achieve species discrimination within some groups. This is particularly the case for the smallest species (< 20 μm), especially those that are delicate, naked, or thin-walled, which are also often overlooked. However, the massive sequencing of environmental samples conducted during the last two decades around the world has clearly shown that the richness of dinoflagellates has been considerably underestimated, with the largest deficits occurring for species < 20 μm in size (Moon-van der Staay et al., Citation2001; Le Bescot et al., Citation2016). Accordingly, the diversity, distribution, abundance, and ecological importance of dinoflagellates merits further, detailed studies.
The genus Ansanella H.J.Jeong, S.H.Jang, Moestrup & N.S.Kang, erected by Jeong et al. (Citation2014), was classified within the family Suessiaceae of the order Suessiales. This assignment was made based on the presence in member species of a type E eyespot (i.e. a series of cisternae with brick-shaped contents), an apical apparatus consisting of an elongated apical vesicle (EAV), and the results of molecular analyses. Jeong et al. (Citation2014) found a larger number of the latitudinal series of amphiesmal vesicles (AVs) in the type species A. granifera H.J.Jeong, S.H.Jang, Moestrup & N.S.Kang than in phylogenetically related genera, such as Pelagodinium, Polarella, Protodinium and Symbiodinium. Other morphological characters were shown to distinguish Ansanella from genera containing a comparable number of horizontal rows of AVs, such as Biecheleriopsis and Biecheleria. Jeong et al. (Citation2014) considered the presence of grana-like thylakoids, observed in A. granifera, as a trait of the Ansanella genus. Recently, a second species was included in the genus, A. natalensis (T.Horiguchi & R.N.Pienaar) Dawut, Sym & T.Horiguchi 2018, transferred from the genus Gymnodinium based on the results of phylogenetic and morphological analyses. Dawut et al. (Citation2018) found that this species shared numerous morphological traits with A. granifera, including the arrangement of its AVs in a similar number of latitudinal rows, the shape and position of the EAV, the presence of a type E eyespot, a similar type of pyrenoid, the absence of a peduncle and nuclear chambers, the lack of a nuclear fibrous connective (NFC), and no resting-cyst-like cells. The authors distinguished the species from A. granifera based on the possession of a single chloroplast rather than the five to eight chloroplasts of A. granifera, the absence of grana-like thylakoids in the chloroplast, and the presence of mucocysts. In addition, the two species were found in two distinct habitats ().
Table 1. Comparison of Ansanella catalana, other Ansanella species and closely related taxa
In this study, we isolated several clonal cultures of small thin-walled dinoflagellates from the Catalan coast. Phylogenetic analyses of the strains allowed their classification as Ansanella members. While some corresponded to A. cf. natalensis, others represented a new, previously unknown species. This species, including its thus far unique morphological features, is described herein together with phylogenetic information for the order Suessiales that will facilitate the identification of the species. Our study also included an exploration of the metabarcoding data available from three different habitats (harbours, beaches and a coastal lagoon) along the Catalan coast, to determine the distribution of Ansanella and the relevance of this genus to coastal dinoflagellate communities.
Materials and methods
Strain isolation and cultures
Clonal cultures of Ansanella representatives were isolated in 2018 from different harbours located along the Catalan coast (Spain), NW Mediterranean Sea (Supplementary table S1). Approximately 2 l of surface water was collected with a bucket and pre-filtered through a 60 µm mesh. Organisms from the pre-filtered water were concentrated on a 5 µm mesh by gravity and collected in a flask. The cells were then observed under an inverted microscope and micropipetted into different wells of a plate. The resulting isolates were grown in K/5 medium (Keller et al., Citation1987) prepared with coastal seawater and with a salinity of 36. The cultures were incubated at 20°C with a 12:12 light:dark (L:D) photoperiod and an irradiance of ~100 µmol photons m−2 s−1.
During the samplings, water temperature and salinity were measured in situ using a microprocessor conductivity meter (YSI model 30M/10 FT) and 60 ml water samples, not pre-filtered, were immediately frozen (−20°C) until nutrient quantification. The concentrations of dissolved inorganic nutrients (NO3, NH4 and PO4) were measured as described by Grasshoff et al. (Citation1983), using an AA3 continuous flow analyser (Bran+Luebbe).
Morphological and ultrastructural analyses
Light microscopy
Cells from ICMB 285 culture were examined and photographed under bright-field and epifluorescence (lamp 50 W) microscopy using a Leica DM IRB (Leica Microsystems GmbH, Wetzlar, Germany) inverted light microscope connected to a ProgRes C10 (JENOPTIK Laser, Optik, Systeme GmbH, Jena, Germany) digital camera and a Zeiss Cell Observer HS microscope. Nuclei were stained with 4’,6-diamidino-2-phenylindole (Sigma-Aldrich, St. Louis, Missouri, USA) at a final concentration of 2 μg ml−1 and photographed through a UV filter. Cellular dimensions were determined in 50 cells using ProgRes capturePro v 2.1 software (JENOPTIK Optical Systems GmbH).
Confocal microscopy
Two ml of a living ICMB 285 culture were used to observe the three-dimensional structure of the dinoflagellate’s chloroplasts. The cells were visualized with a Leica TCS- SP5 II CW-STED inverted confocal microscope system using a HCX PL APO lambda blue 63× 1.40 oil UV objective. The excitation wavelength was 458 nm. Images were processed using Image J.
Scanning electron microscopy (SEM)
ICMB 285 cells were fixed for 15 min at room temperature by the addition of a volume of 4% osmium tetroxide (dissolved in seawater) sufficient to reach a final concentration of 1%. The pH was not adjusted. The fixed cells were then filtered through a 13 mm diameter Nucleopore (Pleasanton, California, USA) polycarbonate filter with a pore size of 3 µm. The filtered cells were washed in distilled water, dehydrated in an ethanol series (25, 50, 75, 96, and 3 × 100%, 10 min each), and critical-point dried in a Bal-Tec CPD030 apparatus (Leica Microsystems, Vienna, Austria). The filters were mounted on stubs, sputter-coated with gold in a Quorum Q150R S (Quorum Technologies, Ltd, Laughton, UK), and examined using a variable-pressure scanning electron microscope (Hitachi S-3500N, Hitachi High Technologies Corporation, Japan) at the Servei de Microscòpia Electrònica (ICM-CSIC).
Transmission electron microscopy (TEM)
For TEM, an aliquot of the ICMB 285 culture was transferred to a 15 ml centrifuge tube and fixed in 2% glutaraldehyde (final concentration) made up with seawater at room temperature. After 4 h, the cells were concentrated by centrifugation for 10 min at 2000 rpm and then transferred to a 1.5 ml tube. The cells in the pellet were rinsed several times in filtered seawater, post-fixed for 90 min in 1% osmium tetroxide (final concentration) prepared in distilled water, pelleted by centrifugation, and then washed in culture medium. The material was dehydrated in an acetone series (50, 70, 80, 3 × 90, 3 × 96, 3 × 100%), embedded in Spurr’s resin, and then sectioned on a Reichert Jung Ultracut E (Capovani Brothers Inc., Scotia, New York, USA) ultramicrotome using a diamond knife (Diatome, Hatfield, Pennsylvania, USA). The sections were collected on a copper 200-mesh grid without film and then stained in 2% uranyl acetate and lead citrate following the method of Reynolds (Citation1963). The sections were examined in a JEM-1010 electron microscope (JEOL USA Inc.) operated at 80 kV. Micrographs were taken using a Gatan, Orius SC 1000 A1 (Gatan Inc., Pleasanton, California, USA) digital camera.
DNA extraction, PCR amplification and phylogenetic analyses
Genomic DNA was extracted from 15–20 ml of each culture using the DNeasy blood & tissue extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted DNA was used as the template to obtain SSU and LSU rDNA sequences. For the SSU rDNA, 1–2 µl of extract was subjected to PCR using the EK-82F–18S-1520R primers. The 25 µl PCR mixture contained 2.5 µl of 10× buffer, 1.5 mM MgCl2, 1.25 U of Platinum Taq DNA polymerase (Invitrogen, Thermo Fisher Scientific Corp.), 0.2 mM of each dNTP and 0.4 mM of each primer. The reaction conditions were as follows: initial denaturation for 2 min at 94°C, 35 cycles of 15 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by a final extension step for 5 min at 72°C. In some cases, a semi-nested PCR was performed to increase the PCR product yield, using 1 µl of the previous PCR product as template, primers EK-82F and 1209R, and the above-described reaction conditions. For LSU rDNA, the PCRs were performed using primers D1R and D3C and the conditions described above. Four µl of each PCR product was electrophoresed in an agarose gel and then visualized under UV illumination. Purification and Sanger sequencing were carried out by external services (Genewiz, Takeley, UK) using both forward and reverse primers. The obtained sequences are listed in Supplementary table S1.
Phylogenetic trees were constructed using all of the Ansanella sequences obtained in this study together with representative sequences of Suessiales available in GenBank. Tovelliaceae and Borghiellaceae representatives were used as outgroups for the LSU rDNA phylogenetic tree and Borghiellaceae representatives for the SSU and concatenated SSU-LSU rDNA phylogenetic trees. For both rDNA regions, the sequences were aligned using the MAFFT v.7 online server (Katoh et al., Citation2002) under the ‘auto’ option. The alignment was manually checked with BioEdit v. 7.0.9 (Hall, Citation1999). Subsequently, the alignments were trimmed using Gblocks with less stringent options (Castresana, Citation2000) to remove poorly aligned regions, resulting in a final alignment of 1690 and 854 positions for the SSU and LSU rDNA regions, respectively. Additionally, Suessiales strains with available SSU and LSU rDNA sequences were selected to construct a concatenated phylogenetic tree (Supplementary table S2). The sequences were aligned and trimmed as explained above. Maximum-likelihood (ML) phylogenetic relationships were determined with RAxML v8.0 (Stamatakis, Citation2014), using GTRGAMMA model and 1000 runs with distinct starting trees. The bootstrap ML analysis was done with 1000 pseudo-replicates. Bayesian inference was run using MrBayes v3.2.1 (Ronquist et al., Citation2012) with a GTR model and four Markov chains with one million cycles for each chain. The Bayesian posterior probabilities were obtained from the post-burn-in consensus tree.
Ansanella distribution
The distribution of Ansanella was explored from a publicly available V4 18S rDNA metabarcoding dataset (NCBI SRA, bioproject number PRJNA630546), based on analyses conducted during 2018 and 2019 at 15 sampling locations from three types of habitat (harbours, beaches and one lagoon) distributed along the Catalan coast. Water temperature and salinity were measured in situ using a microprocessor conductivity meter (WTW model Cond 3301). Nutrient samples were analysed following established procedures. Sediments and seawater representing the depth of the water column were sampled for each location. Seawater was processed by serial filtration, resulting in small (0.8–10 µm) and large (10–200 µm) sample fractions. Sediments were processed independently. Details of the sampling, sample process and data treatment to obtain the final ASVs (amplicon sequence variants) can be found in Reñé et al. (Citation2021). All available Ansanella sequences, including those from this study and from GenBank, were queried for sequence similarity using blastn (Altschul et al., Citation1990) with default parameters against all ASVs. Only sequences with an e-value of 10−5, 100% coverage (i.e. 359 bp), and >99% identity were selected for the subsequent analyses.
Results
Ansanella new strains
Six clonal strains of small dinoflagellates isolated in four different harbours of the Catalan coast were identified based on their rDNA sequences as belonging to the genus Ansanella (see below). The new isolates consisted of four strains phylogenetically very close to A. natalensis and two strains of a new species (Supplementary table S1). The strains closely related to A. natalensis were examined under light microscopy and their morphology agreed with that described in previous studies of the species, including the cell size and number of pyrenoids (Dawut et al., Citation2018), and with the species referred to in this work as A. cf. natalensis. A detailed morphological and ultrastructure characterization of strain ICMB285 revealed it to be a new Ansanella species, named Ansanella catalana. This culture strain was deposited in the Culture Collection of Microalgae of the Instituto Español de Oceanografia and in the Banco Español de Algas under the code BEA 1919B.
Ansanella catalana N.Sampedro, J.Matos & A.Reñé sp. nov. ()
Figs 1–8. Light micrographs of Ansanella catalana sp. nov. Fig. 1. Ventral view showing the eyespot (arrowhead) and the thumb-like protusion (arrows). Fig. 2. A cell containing different pyrenoids (arrows) and the nucleus (n). Fig. 3. Cell showing the elongated apical vesicle (arrow). Fig. 4. Fluorescent micrographs showing the position of the nucleus. Figs 5–8. Confocal microscopy of serial sections of the same cell (from left to right), showing the lobules of the chloroplast and their connections. Scale bars = 5 µm.
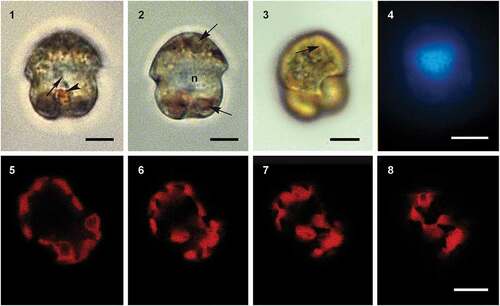
Figs 9–12. Scanning electron micrographs of cells of Ansanella catalana sp. nov. culture strain ICMB 285 showing the rows of amphiesmal vesicles in (Fig. 9) ventral, (Fig. 10) dorsal, (Fig. 11) antapical and (Fig. 12) apical views. E = episome; C = cingulum; H = hyposome; EAV = elongated apical vesicle. Arrowhead points to the small antapical vesicle. Scale bars = 5 µm.
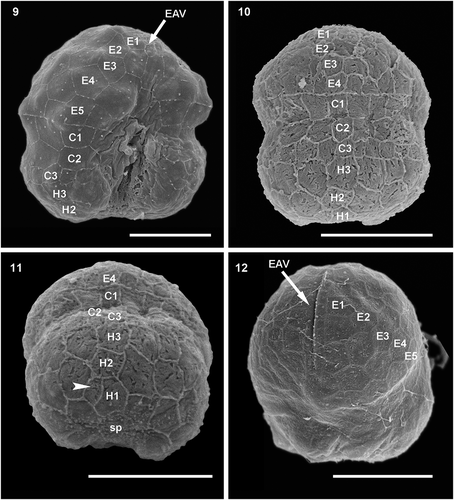
Figs 13–14. Schematic drawing of the amphiesmal vesicles of Ansanella catalana sp. nov. (Fig. 13) Ventral view and (Fig. 14) dorsal view. Scale bars = 2 µm.
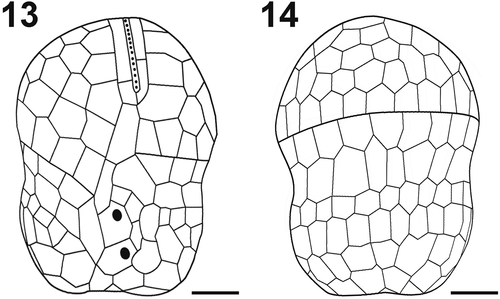
Figs 15–25. Scanning electron micrographs and drawings showing morphological details of cultured A. catalana cells. Fig. 15. Ventral view of the episome reveals the frequently seen elongated AV in contact with the EAV (asterisk) and the consistently detected Sda. Fig. 16. Hyposome and the three rows of AVs. Fig. 17. Apical view of the EAV. Fig. 18. Drawing of the EAV highlights the long central AV ornamented with knobs and the surrounding platelets. Fig. 19. Cingulum of a cell showing the zig-zag line of its lower margin. Fig. 20. EAV and its knobs (arrowhead). Fig. 21. Drawing of the sulcal area. Fig. 22. Sulcal area of a cell, showing a thumb-like protrusion (double arrowhead). Fig. 23. Possible resting cyst. Figs 24, 25. Dividing cells showing oblique division. E = episome; C = cingulum; H = hyposome; EAV = elongated apical vesicle; Sa = anterior sulcal vesicle; Sda = right anterior sulcal AV; Sp = posterior sulcal vesicle; Scale bars=1 µm (Figs 15, 20, 21), 2 µm (Fig. 16), 3 µm (Figs 17, 19, 22) and 5 µm (Figs 23, 24, 25).
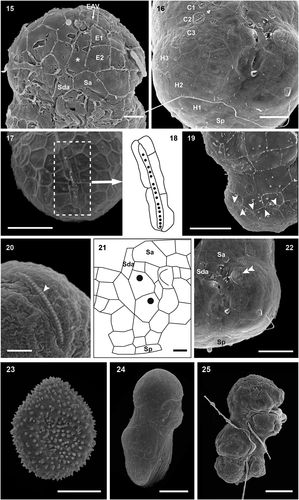
Figs 26–29. Transmission electron micrographs of cells of Ansanella catalana sp. nov. in longitudinal sections (Figs 26, 27) and transverse sections at different planes of the cells (Figs 28, 29). Several organelles can be seen: chloroplasts (c), eyespot (e), mitochondria (m), nucleus (n), and pyrenoid (py). Scale bars = 2 µm.
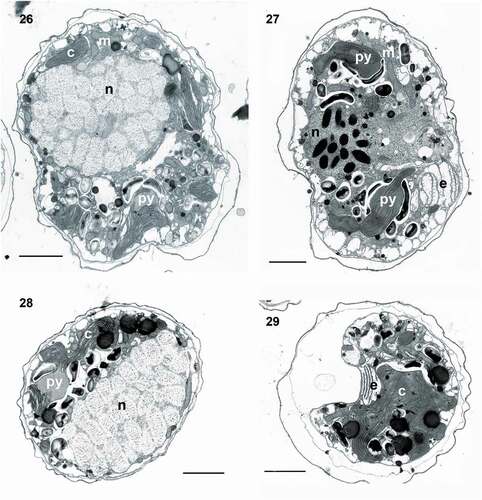
Figs 30–36. Transmission electron micrographs of cells of Ansanella catalana sp. nov. (Fig. 30) Detail of the nucleus showing the nucleolus (nu) and Golgi apparatus (g). Fig. 31. Detail of the fibrous vesicle (f) located close to the nucleus(n). Fig. 32. The chloroplast (c), containing > 3 stacked thylakoids per lamella. Fig. 33. Cell containing a long fibrous vesicle and pyrenoids (py) penetrated by thylakoids, one of them behind the eyespot (e). Fig. 34. Detail of a chloroplast containing three thylakoids stacked per lamella. Fig. 35. Chloroplast with connected lobes. Fig. 36. Pyrenoid embedded in the chloroplast matrix and surrounded by starch sheets (s). Scale bars = 2 µm (Figs 30, 31, 33, 35), 200 nm (Figs 32, 34), and 1 µm (Fig. 36).
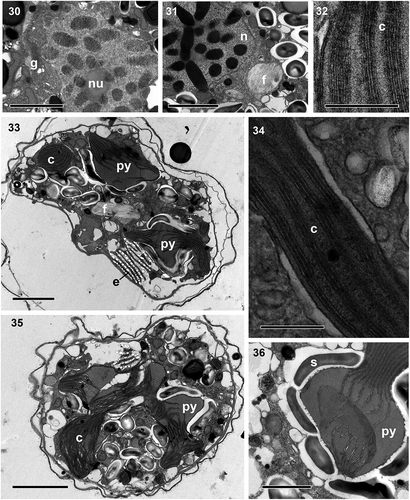
Figs 37–44. Transmission electron micrographs. Fig. 37. Numerous extrusomes, some of them with lid structures (double arrowhead), are seen in the peripheral part of the cell. Figs 38–39. Details of an extrusome, comprising an anterior chamber (ac) filled with diffuse material and a posterior chamber (pc) with electron-denser material with a striated pattern. Fig. 40. Transverse section of the anterior (ac) and posterior (pc) chambers. Fig. 41. Transverse section of the posterior chamber. Fig. 42. Globular mucocyst (mu). Fig. 43. Detail of the amphiesma, consisting of an inner amphiesmal membrane (IAM), pellicle (P), outer amphiesmal membrane (OAM), and plasma membrane (PM). Fig. 44. Detail of the eyespot (e), shows its cisternae containing brick-like material and a pyrenoid (py) situated behind it. Scale bars = 1 µm (Figs 37, 43, 44), 500 nm (Figs 38, 39, 42) and 100 nm (Figs 40, 41).
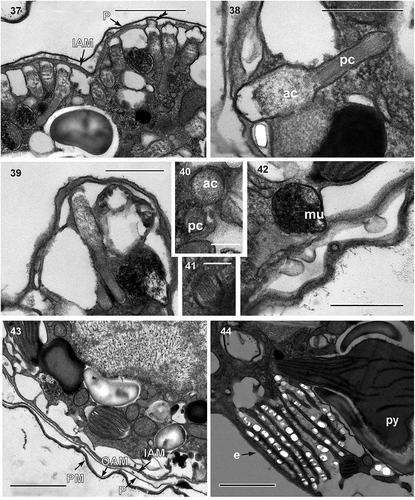
Figs 45–47. Fig. 45. Cell showing a probable pusule system; Fig. 46. putative pusule (pu); Fig. 47. putative common pusule canals (cpc). Scale bar = 5 µm (Fig. 45), 1 µm (Figs 46, 47).
DIAGNOSIS: Free-living photosynthetic dinoflagellate, 9.6–15.5 μm in length (L), 7.3–11.8 μm in width (W), and a L/W ratio of 1.2–1.6 μm. Cells with an almost hemispherical episome and a truncated hyposome narrower and shorter than the episome. The descending cingulum is displaced by an amount almost equal to its own width. Cell surface covered with thin polygonal amphiesmal vesicles arranged in 10–13 latitudinal rows, 4–6 in the episome, 3 in the cingulum and 3–4 in the hyposome. Cell apex with an apical furrow apparatus comprising a single EAV ornamented with knobs. Nucleus located in the central to anterior part of the cell. Single chloroplast with lobules arranged mainly around the cell and containing 2–5 pyrenoids penetrated by thylakoids and surrounded by starch sheets. Type E eyespot present at the sulcal region. Mucocysts and elongated extrusomes consisting of two chambers present.
HOLOTYPE: , obtained from culture strain ICMB285. The SEM stub containing the holotype specimen is deposited in the Biological Reference Collections (CBR) at the Institut de Ciències del Mar (ICM-CSIC, Barcelona, Spain) under the accession number ICMCBR000462 (Guerrero et al., Citation2020).
GENBANK ACCESSION NUMBERS: SSU rDNA: MT432078; LSU rDNA: MT432072.
TYPE LOCALITY: Olympic Harbour (Port Olímpic), Barcelona, Catalonia, NW Mediterranean Sea (41°23.074′N, 2°12.001′E).
ETYMOLOGY: Named after the Catalan coast, where it was first detected.
DISTRIBUTION: NW Mediterranean Sea.
HABITAT AND ECOLOGY: Marine coastal phytoplankton. Strain ICMB285 was isolated in May from Olympic Harbour (Port Olímpic) at a water temperature of 18.8°C and a salinity of 35.4. Strain ICMB284 was isolated in February from Arenys Harbour at a water temperature of 12.4°C and a salinity of 37.1. Nitrate, ammonium and phosphate concentrations in Arenys Harbour were 4.11 µmol l–1, 0.45 µmol l–1 and 0.19 µmol l–1, respectively, indicating the ability of this species to grow under eutrophic conditions over a temperature range of at least 12.4–18.8°C.
Swimming behaviour: Cells swim in straight lines or in loops in different directions, but they also produce quick straight movements, similar to short jumps, usually after a small loop.
Cell morphology: The episome is rounded, forming almost a hemisphere, while the hyposome is slightly narrower and shorter than the episome, with a truncated to bilobate end (). The wide descending cingulum is displaced by a distance almost equal to its own width (, ). Living cells range in length (L) from 9.6–15. 5 μm (average 12.5 ± 1.5 μm; n = 50) and in width (W) from 7.3–11.8 μm (average 9.4 ± 1.2 μm; n = 50), resulting in elongated cells with a L/W ratio of 1.2–1.6 μm (average 1.3 ± 0.1 μm; n = 50). The longitudinal flagellum is 14–16 μm long and ~345 nm thick. The nucleus is large and centrally located (). The yellowish-brown chloroplast lobules are arranged around the periphery of the cell and contain 2–5 round pyrenoids (). A bright reddish-orange eyespot is located in the sulcal area ().
The cell surface is covered with small polygonal (quadrangular, pentagonal or hexagonal) AVs measuring 1–1.6 μm in length and arranged in 10–13 latitudinal rows, with 4–6 latitudinal rows belonging to the episome, 3 to the cingulum, and 3–4 to the hyposome (, ). While the upper border of the cingular area is clearly delimited, forming a straight line with the lower episome row, this is not the case for the lower border (). The AVs of row C3 are smaller than those of the other cingular rows and form an irregular, zigzag line with those of the hyposomal row ().
The AV situated below the EAV in ventral view is mostly elongated towards the sulcus (). In some specimens two AVs may be present instead of a single elongated AV (not shown). This ventral and elongated AV is usually in contact with three AVs on the left side. The uppermost of these three AVs is also in contact with the EAV, with the bottom part of the middle AV bordering the cingulum and the lowermost AV equivalent to the anterior sulcal plate (Sa). The latter is pentagonal in shape and is bordered on the left side by the first cingular AV, along a diagonal edge (). The posterior part of the elongated vesicle is in contact with another elongated, narrower vesicle, the right anterior sulcal AV (, , ). The sulcus widens towards the antapex and seems to be made up of 7–8 rows of AVs (). A protruding, thumb-like AV is present in the hyposome, in the left corner of the intercingular-sulcal region (, ). The hyposome may permanently feature a small antapical vesicle (). The episome forms an apical furrow complex with a single EAV ~5 μm in length and including a central line made up of 26–27 small knobs surrounded by 5–8 platelets (, , , , ). No peduncle was observed emerging from the flagellar area.
Life cycle features: A possible resting cyst of Ansanella catalana was observed by SEM. The cyst had a conical episome, a rounded hyposome, and measured ~12 μm × 10 μm (L×W). The surface of the cyst was ornamented with tiny conical spinules ~50 nm in length (). Certain resting stages were not observed in the cultures examined by light microscopy. Dividing cells were observed surrounded by a pellicle layer (). Cell division was oblique () and during division, the cingula of the two daughter cells remained parallel to each other.
Cell ultrastructure: The nucleus of A. catalana is located in the central to anterior part of the cell, occupying approximately half of the length of the cell at its dorsal end (). Numerous chromosomes and a nucleolus () are present in the nucleus; the nuclear envelope features nuclear pores (not shown) but not nuclear chambers. None of the observed cells seemed to contain an NFC, but they were not examined in serial sections. The Golgi apparatus and fibrous vesicles are located close to the nucleus (). The lobules of a single chloroplast are arranged around the cell (, , ). Each lamella typically possesses three thylakoids (), but arrangements of a larger number of thylakoids stacked per lamella are also observed (). The chloroplast contains 2–5 pyrenoids, also visible under light microscopy (, , , ), with one of them typically situated behind the eyespot (, ). The pyrenoids are embedded in the chloroplast matrix, penetrated by thylakoids and surrounded by starch sheets. Numerous starch grains and mitochondria are present in the central and peripheral parts of the cell, and numerous extrusomes of two different types in the periphery: (1) a mucocyst made up of a globular or oval vesicle with a dense granular content and measuring ~350 nm in diameter () and (2) a more complex extrusome consisting of two chambers of approximately the same length (). The anterior chamber is wider than the posterior chamber, circular in cross-section, and contains diffuse material (). It is usually truncated in its distal part, which is in contact with the inner amphiesmal membrane, and positioned just under the apical lid structure, which is located in the AV (). The posterior chamber contains a core of electron-dense material with a striated pattern. It is square in transverse section () and with a rounded end (). This elongated extrusome measures ~1.1 µm in length, with the anterior chamber having a width of 190–250 nm and the posterior chamber a width of 120–140 nm. A type E eyespot (a stack of cisternae containing brick-like material) is present in the hyposome, underneath the sulcus (). It consists of ~8 cisternae, each forming a line of brick-like structures enclosed in a matrix (). The eyespot is ~2 µm in width and 2–2.5 µm in length. Components probably belonging to the pusule system are also observed close to the sulcus ().
Molecular and phylogenetic analyses: SSU rDNA sequences obtained for strains ICMB 284 and ICMB 285, corresponding to A. catalana, were identical, as were those obtained for ICMB 286, ICMB 287 and ICMB 289, corresponding to A. cf. natalensis. The latter had a 99.7% similarity with A. natalensis sequences available from South Africa. Sequences of A. catalana had a 99.8% similarity with sequences of A. granifera, a 99.8% similarity with the A. cf. natalensis sequences from this study, and a 99.6% similarity with sequences from South Africa.
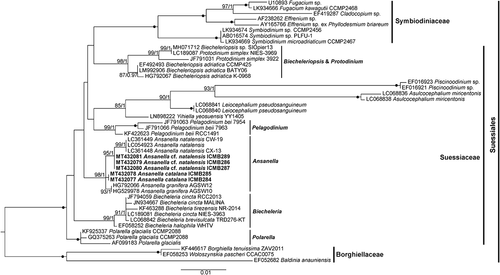
According to the ML phylogenetic tree based on the SSU rDNA sequences (), maximum support was obtained for the Suessiales clade, which included representatives of the family Symbiodiniaceae (100%/1) and the genera Biecheleriopsis and Protodinium (98%/1), Pelagodinium (99%/1), Biecheleria (99%/1), Polarella (100%/1) and Ansanella (98%/1). However, the phylogenetic relationships of the different genera were completely unresolved. Other monotypic genera, such as Yihiella, Leiocephalium, Asulcocephalium and Piscinoodinium, clustered together (85%/1). Within the Ansanella clade, A. granifera sequences clustered together (93%/1). A. cf. natalensis also formed an independent clade (99%/1), but the sequences from South Africa clustered together (99%/1). Finally, A. catalana sequences formed an independent clade, but with no statistical support.
Fig. 48. Maximum likelihood phylogenetic tree inferred from SSU rDNA sequences, including Suessiales representatives. Sequences of Borghiellaceae species were used as the outgroup. Sequences from this study are highlighted in bold. Bootstrap (BS) values and Bayesian posterior probabilities (BPP) are provided (% BS/BPP) only when > 80% and > 0.9, respectively.
The LSU rDNA sequences obtained for A. catalana were identical, as were those obtained for A. cf. natalensis. The latter were 99.8% similar to the sequences available in GenBank. A. catalana sequences had a 98.2% similarity with those of A. natalensis, a 98.4% similarity with those of A. cf. natalensis, and a 98.4% similarity to those of A. granifera. As in previous studies of Ansanella representatives (Jeong et al., Citation2014; Dawut et al., Citation2018), the generated LSU rDNA sequences did not show a 51-base pair deletion in the D2 domain, in contrast to other Suessiaceae genera such as Biecheleria, Yihiella, Leiocephalium and Asulcocephalium (; Jang et al., Citation2017).
In the ML phylogenetic tree based on LSU rDNA sequences, Ansanella representatives did not form a monophyletic clade, while in the Bayesian reconstruction they clustered together but were characterized by polytomy (Supplementary fig. S1). All Suessiales representatives formed a highly supported clade (96%/1), including Symbiodiniaceae (100%/1), Biecheleriopsis and Protodinium (100%/1), Biecheleria (98%/1), Pelagodinium (84%/1), Polarella (100%/1) and Ansanella representatives. Other monotypic genera, such as Dactylodinium, Asulcocephalium, Leiocephalium and Yihiella, clustered independently at the base of the clade. As observed for the SSU rDNA phylogeny, the phylogenetic relationships between all genera were mostly unresolved. For Ansanella representatives, sequences belonging to A. cf. natalensis and A. natalensis clustered together (99%/0.92), as did those belonging to A. catalana (99%/0.96). Both formed sister clades with the sequence of A. granifera.
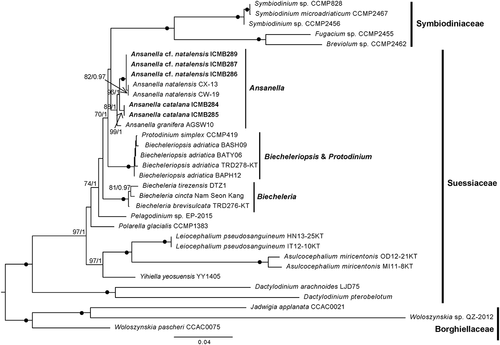
The SSU-LSU rDNA concatenated phylogenetic tree () confirmed the results obtained for the single-gene phylogenies. All Ansanella representatives formed a moderately to highly supported clade (88%/1). Sequences corresponding to A. cf. natalensis and A. natalensis clustered together (100%/1), but the latter formed a cluster (82%/0.97) independent of the sequences obtained in this study. Sequences of A. catalana formed an independent clade (99%/1), and the sequence of A. granifera clustered at the base of the clade. As noted above, clusters with maximum support were obtained for the different genera of Suessiaceae. In this case, internal nodes had higher statistical supports than was the case in the single-gene phylogenies. A clade comprising Ansanella, Symbiodiniaceae and Biecheleriopsis representatives was obtained with moderate support (70%/1).
Fig. 49. Maximum likelihood phylogenetic tree inferred from the concatenated SSU + LSU rDNA sequences, including Suessiales representatives. Sequences of Borghiellaceae species were used as the outgroup. Sequences from this study are highlighted in bold. BS values and BPP are provided (% BS/BPP) only when > 70% and > 0.9, respectively.
Ansanella distribution, relative abundances and environmental conditions along the Catalan coast
Representatives of Ansanella were found in the metabarcoding dataset, and four ASVs matched the Ansanella sequences. As previously shown, the sequences obtained for A. cf. natalensis slightly differed from the South African sequences available in GenBank. One ASV (ASV5) had a 100% match with sequences of both A. catalana and A. cf. natalensis from this study but this ruled out a species distinction based on the V4 region. A second ASV (ASV401) had a 100% match with sequences of A. natalensis from South Africa. Finally, two ASVs (ASV3228 and 3229) that diverged from the sequences of A. catalana at 2 and 3 positions, respectively, were detected. ASV5 was present at all sampling locations except in Barcelona Harbour (), and with a few exceptions in both (small and large) fractions and in the sediments. However, its relative abundance was generally higher in the small (0.8–10 µm) fraction. ASV5 was detected in the water collected from nine beaches, with relative abundances ranging from 0.05% at Cubelles to 61.5% at L’Arenal. The relative abundances detected in sediments were much lower, with a maximum of 3.2% at L’Arenal. In the harbours, the abundance of ASV5 was <4% except in Vilanova Harbour, where it accounted for 24.6% of the dinoflagellate community. In the sediments, the abundances were also lower, with a maximum of 2.1% at Vilanova Harbour. Finally, in the samples from the single coastal lagoon included in the dataset, ASV5 contributed 97% of the dinoflagellate sequences in the small fraction, 64.4% of those in the large fraction and 11% of those in the sediments.
Fig. 50. Distribution and relative abundances of Ansanella (ASV5) in the dinoflagellat e communities from the water column (2 fractions) and sediments. The sampled area included three different coastal habitats along the Catalan coast (NE of Spain). Missing symbols in some locations represent no detection in that fraction.
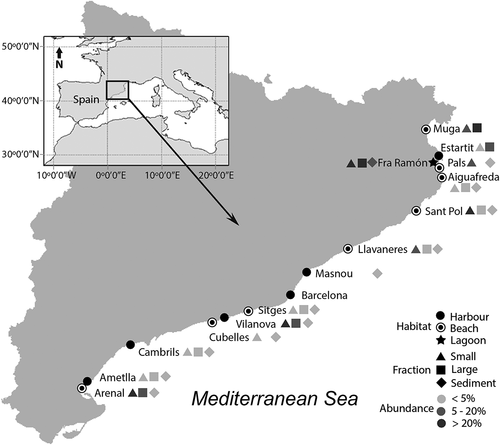
ASV401, corresponding to A. natalensis (South Africa strain), was detected in both the large (10–80 µm) fraction and the sediments of L’Estartit Harbour as well as in the sediments of Vilanova Harbour, representing 1.2%, 1.4% and 2.3% of the dinoflagellate community, respectively. Both ASV3228 and ASV3229 were detected only in the small fraction from the Fra Ramon coastal lagoon, with identical read numbers and each representing 0.2% of the dinoflagellates community sequences.
Our results on the environmental conditions that host Ansanella showed that members of this genus tolerate a wide range of salinities (30.5–52.6), temperatures (11.8–29°C) and inorganic nutrient levels, including those of nitrate (0.03–8.66 μmol l–1), ammonium (0.26–11.29 μmol l–1) and phosphate (0.04–2.95 μmol l–1). The highest relative abundances were detected at high water temperature (> 21.5°C) but at the two ends of the salinity range.
Discussion
Ansanella catalana shows the typical characters of the Suessiaceae family: a type E eyespot, an apical structure formed by an EAV, and AVs organized in > 7 latitudinal rows (Moestrup et al., Citation2009b). The phylogenetic analyses performed in this study showed the close relationships of the new isolates and Suessiaceae genera, such as Biecheleriopsis, Biecheleria and Protodinium, and particularly with members of Ansanella.
The results of our molecular analyses also indicated that our new species is related to A. granifera and A. natalensis. Like the latter two species, A. catalana has the typical characters of the Suessiaceae described above, but also a zigzag pattern of the lower margin of the cingulum, the absence of an NFC, peduncle and nuclear chambers, and a similar number of latitudinal rows. Together, these features clearly indicate that our newly identified species belongs to the genus Ansanella. Moreover, its description allows for a more robust delimitation of the genus from the other related genera (). For example, the genus Biecheleriopsis, described in previous studies (), differs from Ansanella by the straight pattern of the lower margin of the cingulum, the presence of a NFC, and thylakoids with swollen terminal ends (Moestrup et al., Citation2009b). The genus Protodinium, possibly congeneric with Biecheleriopsis (Luo et al., Citation2015), differs from Ansanella by the number of latitudinal rows of the episome and the morphology of the lower margin of the cingulum (). The presence of a peduncle, the lack of a 51-base pair fragment in domain D2 of the large subunit of its rDNA, and the formation of spherical resting cysts with small spines or thin hair-like bristles are features that distinguish Biecheleria from Ansanella (Moestrup et al., Citation2009a; Jeong et al., Citation2014; Takahashi et al., Citation2014; ).
Other features have been proposed to distinguish Ansanella from related genera. The possession of grana-like thylakoids was suggested by Jeong et al. (Citation2014) as a taxonomic character limited to Ansanella, but a similar structure was observed in Leiocephalium pseudosanguineum of the Suessiaceae (Takahashi et al., Citation2015) and not observed in A. natalensis (Dawut et al., Citation2018). Based on the latter, it was discarded as an identifying character. The absence of resting-cyst formation was cautiously invoked as a character of Ansanella representatives (Dawut et al., Citation2018), but in this study a putative resting cyst was observed, similar to the cysts described for Yihiella yeosuensis (Jang et al., Citation2017) and for Biecheleriopsis adriatica by Moestrup et al. (Citation2009b). Cysts of B. adriatica are rarely observed (Moestrup et al., Citation2009a; Kang & Wang, Citation2018), which may also be the case for A. catalana and other Ansanella species. The detection of Ansanella sequences in sediments from almost all sampling locations suggests that resting stages are part of the life cycle of this genus. However, because sediments were obtained in shallow environments, that the obtained sequences corresponded to planktonic organisms present on the sediment surface cannot be excluded. However, Ansanella sequences were also detected in sediments but not in the water samples from El Masnou Harbour. Thus, the absence of resting cyst cannot be used to distinguish Ansanella from other related genera.
Despite the polytomy of Ansanella representatives in the phylogenetic tree inferred from LSU rDNA sequences, the concatenated SSU-LSU rDNA phylogenetic tree showed a well-supported clade of Ansanella species in which A. catalana formed an independent and well-supported clade. This relationship is consistent with the morphological and ultrastructural characters of this new species. A. catalana differs from the other two Ansanella species recognized to date by morphological details of the cell surface and the presence of two types of extrusomes, whereas extrusomes were not reported for A. granifera and only mucocysts were reported for A. natalensis (). Furthermore, A. catalana possesses a single chloroplast, in contrast to A. granifera, although this feature is difficult to observe without confocal microscopy because of the complex ramified structure that makes up the chloroplast. Externally, A. catalana shows a small AV in the antapical part of the cell, which is not observed in either A. granifera or A. natalensis. In addition, A. catalana usually has only one elongated AV, located on the ventral part of the episome between the EAV and the right anterior sulcal AV, in contrast to the two or three AVs observed in A. granifera and A. natalensis, respectively (). However, other characters previously considered to delimit Ansanella species are not valid. In the above-cited study by Dawut et al. (Citation2018), the presence of grana was discarded as an Ansanella character but it was still used to distinguish A. granifera from A. natalensis. However, an earlier study pointed out that the presence of grana is not useful as a specific taxonomic character because it differs between strains of the same species (Boutrup et al., Citation2017). Similarly, our metabarcoding results showed that V4 18S rDNA is not a reliable marker to distinguish among Ansanella species, since it failed to discriminate between A. catalana and A. cf. natalensis (Catalonia strains).
Extrusomes
To the authors’ knowledge, the extrusome found in A. catalana, resembling a combination of a trichocyst and a mucocyst, has not been observed in other dinoflagellates. Rhiel (Citation2017) compiled a list of the extrusomes seen in dinoflagellates (table 1 in Rhiel, Citation2017), but other types have been observed in Suessiales species such as Yihiella yeosuensis (Jang et al., Citation2017), Dactylodinium pterobelotum (Takahashi et al., Citation2017) and D. arachnoides (Lum et al., Citation2019). The trichocyst with lateral hairs found in Dactylodinium species (Takahashi et al., Citation2017; Lum et al., Citation2019) is similar to the extrusome found in A. catalana and also has two well-defined chambers. The posterior chamber in all of these species has a fibrous crystalline core and is square in transverse section. However, while the anterior chamber of the Dactylodinium trichocyst has a well-defined structure made up of fibres and lateral hairs, the extrusome of A. catalana contains diffuse material. Furthermore, the shape of this chamber differs between the extrusomes of the two taxa. In A. catalana, the distal area of the anterior chamber is truncated, while in D. pterobelotum and D. arachnoides it has a conical shape (fig. 6 in Takahashi et al., Citation2017; in Lum et al., Citation2019). Although D. pterobelotum was assigned by Takahashi et al. (Citation2017) to the family Borghiellaceae on the basis of its morphological features, particularly the structures of the eyespot and apical complex, its phylogenetic position as inferred from its LSU rDNA and its SSU-LSU rDNA concatenated is within the Suessiaceae clade (Takahashi et al., Citation2017; ). The authors speculated that some structures of Suessiaceae, such as the eyespot, developed from Borghiellaceae. Similarly, the extrusome of A. catalana may have evolved from the trichocyst of D. pterobelotum or D. arachnoides. However, whether this is the case or the observed extrusome is indeed a new type of extrusome remains to be determined in further studies. Regardless, the clade of Suessiaceae (including Dactylodinium), as a result of the recent incorporation of new species, now contains a remarkable variety of extrusomes (Jang et al., Citation2017; Takahashi et al., Citation2017; Dawut et al., Citation2018; Lum et al., Citation2019; and present paper), including types not previously reported.
With respect to mucocysts, while they have been observed in many species of dinoflagellates their presence in Suessiaceae is thus far limited to two genera, Piscinoodinium (Lom & Schubert, Citation1983; Levy et al., Citation2007) and Ansanella (Dawut et al., Citation2018 and this paper).
Mucocysts consist of vesicles containing diffuse fibrous material, but the mucocysts from different species and even those from the same species may slightly differ in their appearance and structure. Based on the shape, two types of mucocysts, vermiform and flask-shaped, are more frequently reported in dinoflagellates (e.g. Hoppenrath & Leander, Citation2008). The mucocysts observed in A. catalana were flask-shaped, with a spherical to oval body, and measured less than 1 μm. Similar mucocysts, although larger and not spherical, have been observed in A. natalensis (Dawut et al., Citation2018).
Distribution, diversity and ecology of Ansanella genus
To our knowledge, Ansanella was not previously detected in the Mediterranean Sea. However, the metabarcoding results showed that members of this genus are in fact widespread along the Catalan coast and remarkably abundant in the dinoflagellate community. Unfortunately, because A. catalana and A. cf. natalensis (Catalan strains) cannot be distinguished based on their V4 18S rDNA, the specific distribution of each species along the Catalan coast could not be determined.
Dawut et al. (Citation2018) observed that the habitats of A. granifera and A. natalensis were markedly different: while A. granifera was recovered from the estuarine waters of Korea, A. natalensis was found in a tidal pool in South Africa (). Our study adds other habitats for this genus, as we detected A. natalensis in Mediterranean harbours as well as the coexistence of different Ansanella species (Supplementary table S1). The presence of this genus in three different coastal habitats provides evidence of its plasticity. Thus, the Ansanella genus seems to include euryhaline organisms capable of reaching high relative abundances in brackish waters receiving riverine inputs, such as L’Arenal and La Muga, but also in hypersaline environments, such as the Fra Ramón lagoon. However, because the species that formed the ASV5 blooms at these sites could not be distinguished, neither could their habitat preferences.
The differences in the LSU rDNA sequences of A. natalensis and A. cf. natalensis were not significant and the molecular differences in their SSU rDNA may reflect intraspecific variability. However, a V4 SSU rDNA barcode distinct from that of the Catalan A. cf. natalensis strain but coinciding with that of South African A. natalensis has been detected along the Catalan coast. Thus, the strains isolated in this study might represent a new species close to A. natalensis, with the two species coexisting in Catalan waters. Consequently, strains of A. cf. natalensis need to be more carefully examined for morphological differences with A. natalensis. Moreover, other new strains, and thus perhaps new species, were detected by metabarcoding, indicating that the diversity of this genus has yet to be completely described. Without a more accurate description and identification, the distributions and habitats of the different species within this abundant dinoflagellate genus remain elusive.
Supplementary table S1. List of strains obtained in this study, including collection date and place as well as environmental conditions and GenBank accession numbers of sequences obtained.
Supplementary table S2. List of sequences obtained in this study (in bold) and those from GenBank used to construct the concatenated phylogenetic tree, including the corresponding species, strain/isolate and GenBank accession numbers.
Supplementary figure S1. Bayesian phylogenetic tree inferred from the LSU rDNA sequences, including Suessiales representatives. Sequences of Borghiellaceae and Tovelliaceae species were used as the outgroup. Sequences from this study are highlighted in bold. Bootstrap (BS) values and Bayesian posterior probabilities (BPP) are provided (% BS/BPP) only when >80% and >0.9, respectively.
Author contributions
N. Sampedro: sampling, isolation of strains, manuscript concept, light and electron microscopy, morphological analyses, drawings, data presentation, drafting and editing of the manuscript; A. Reñé: sampling, obtaining and processing the sequences, phylogenetic analysis and supporting writing; J. Matos: isolation of strains, maintaining cultures and light microscopy; JM. Fortuño: supporting SEM analysis; E. Garcés: funding acquisition, supervision and supporting writing.
Supplemental Material
Download TIFF Image (63.2 KB)Supplemental Material
Download MS Word (17.1 KB)Supplemental Material
Download MS Word (13.9 KB)Acknowledgements
We thank E. Prats and JM. Rebled from the Unitat de Microscòpia Electrònica, Facultat de Medicina-SCT, Universitat de Barcelona for support with the transmission electron microscopy method, A. Mallabiabarrena for guidance in the confocal microscopy study, and R. Gallisai (ICM-CSIC) and N. Timoneda (ICM-CSIC) for assistance during the molecular and bioinformatic analyses, respectively. This work acknowledges the ‘Severo Ochoa Centre of Excellence’ accreditation (CEX2019-000928-S).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at https://doi.org/10.1080/09670262.2021.1914861
Additional information
Funding
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215: 403–410.
- Benico, G.A., Takahashi, K., Lum, W.M., Yñiguez, A.T., Azanza, R.V., Leong, S.C.Y., Lim, P.T. & Iwataki, M. (2019). First report of Biecheleriopsis adriatica in Bolinao, Northwestern Philippines and its wide distribution in Southeast Asia and adjacent waters. Philippine Journal of Natural Sciences, 24: 34–41.
- Boutrup, P.V., Moestrup, Ø., Tillmann, U. & Daugbjerg, N. (2017). Ultrastructure and phylogeny of Kirithra asteri gen. et sp. nov. (Ceratoperidiniaceae, Dinophyceae) –– a free-living, thin-walled marine photosynthetic dinoflagellate from Argentina. Protist, 168: 586–611.
- Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17: 540–552.
- Dawut, M., Sym, S.D. & Horiguchi, T. (2018). Re-investigation of Gymnodinium natalense (Dinophyceae), a tidal pool dinoflagellate from South Africa and the proposal of a new combination Ansanella natalensis. Phycological Research, 66: 300–309.
- Gómez, F. (2012). A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Systematics and Biodiversity, 10: 267–275.
- Grasshoff, K., Ehrhardt, M. & Kremling, K. (1983). Methods of Seawater Analysis. 2nd ed. Verlag Chemie, Weinheim.
- Guerrero, E., Abelló, P., Lombarte, A., Villanueva, R., Ramón, M., Sabatés, A. & Santos, R. (2020). Biological Reference Collections ICM-CSIC. v1.28. Institute of Marine Sciences (ICM-CSIC). Dataset/Occurrence. https://doi.org/https://doi.org/10.15470/qlqqdx.
- Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95–98.
- Hoppenrath, M. (2017). Dinoflagellate taxonomy – a review and proposal of a revised classification. Marine Biodiversity, 47: 381–403.
- Hoppenrath, M., Elbrächter, M. & Drebes, G. (2009). Marine phytoplankton: selected microphytoplankton species from the North Sea around Helgoland and Sylt. E. Schweizerbart’sche Verlagsbuchhandlung, Stuttgart.
- Hoppenrath, M. & Leander, B.S. (2008). Morphology and molecular phylogeny of a new marine sand-dwelling Prorocentrum species, P. tsawwassenense (Dinophyceae, Prorocentrales), from British Columbia, Canada. Journal of Phycology, 44: 451–466.
- Horiguchi, T. & Pienaar, R.N. (1994). Ultrastructure and ontogeny of a new type of eyespot in dinoflagellates. Protoplasma, 179: 142–150.
- Jang, S.H., Jeong, H.J., Moestrup, Ø., Kang, N.S., Lee, S.Y., Lee, K.H., Lee, M.J. & Noh, J.H. (2015). Morphological, molecular and ecophysiological characterization of the phototrophic dinoflagellate Biecheleriopsis adriatica from Korean coastal waters. European Journal of Phycology, 50: 301–317.
- Jang, S.H., Jeong, H.J., Moestrup, Ø., Kang, N.S., Lee, S.Y., Lee, K.H. & Seong, K.A. (2017). Yihiella yeosuensis gen. et sp. nov. (Suessiaceae, Dinophyceae), a novel dinoflagellate isolated from the coastal waters of Korea. Journal of Phycology, 53: 131–145.
- Jeong, H.J., Jang, S.H., Moestrup, Ø., Kang, N.S., Lee, S.Y., Potvin, É. & Noh, J.H. (2014). Ansanella granifera gen. et sp. nov. (Dinophyceae), a new dinoflagellate from the coastal waters of Korea. ALGAE, 29: 75–99.
- Kang, W. & Wang, Z-H. (2018). Identification of a marine woloszynskioid dinoflagellate Biecheleriopsis adriatica and germination of its cysts from southern Chinese coasts. Journal of Environmental Sciences, 66: 246–254.
- Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30: 3059–3066.
- Keller, M.D., Selvin, R.C., Claus, W. & Guillard, R.R.L. (1987). Media for the culture of oceanic ultraphytoplankton. Journal of Phycology, 23: 633–638.
- Le Bescot, N., Mahé, F., Audic, S., Dimier, C., Garet, M.J., Poulain, J., Wincker, P., de Vargas, C. & Siano, R. (2016). Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environmental Microbiology, 18: 609–626.
- Levy, M.G., Litaker, R.W., Goldstein, R.J., Dykstra, M.J., Vandersea, M.W. & Noga, E.J. (2007). Piscinoodinium, a fish-ectoparasitic dinoflagellate, is a member of the class Dinophyceae, subclass Gymnodiniphycidae: convergent evolution with Amyloodinium. Journal of Parasitology, 93: 1006–1015.
- Lom, J. & Schubert, G. (1983). Ultrastructural study of Piscinoodinium pillulare (Schaperclaus, 1954) Lom, 1981 with special emphasis on its attachment to the fish host. Journal of Fish Diseases, 6: 411–428.
- Lum, W.M., Takahashi, K., Benico, G., Takayama, H. & Iwataki, M. (2019). Dactylodinium arachnoides sp. nov. (Borghiellaceae, Dinophyceae): a new marine dinoflagellate with a loop-shaped apical structure complex and tubular membranous extrusomes. Phycologia, 58: 661–674.
- Luo, Z., Yang, W., Xu, B. & Gu, H. (2013). First record of Biecheleria cincta (Dinophyceae) from Chinese coasts, with morphological and molecular characterization of the strains. Chinese Journal of Oceanology and Limnology, 31: 835–845.
- Luo, Z., Yang, W., Xu, B., Zheng, B. & Gu, H. (2015). Morphology, ultrastructure, and phylogeny of Protodinium simplex and Biecheleriopsis cf. adriatica (Dinophyceae) from the China Sea. Nova Hedwigia, 101: 251–268.
- Moestrup, Ø. & Calado, A.J. (2018). Süßwasserflora von Mitteleuropa, Bd. 6 – Freshwater Flora of Central Europe, Vol. 6: Dinophyceae. Springer, Berlin.
- Moestrup, Ø., Lindberg, K. & Daugbjerg, N. (2009a). Studies on woloszynskioid dinoflagellates IV: the genus Biecheleria gen. nov. Phycological Research, 57: 203–220.
- Moestrup, Ø., Lindberg, K. & Daugbjerg, N. (2009b). Studies on woloszynskioid dinoflagellates V. Ultrastructure of Biecheleriopsis gen. nov., with description of Biecheleriopsis adriatica sp. nov. Phycological Research, 57: 221–237.
- Moon-Van Der Staay, S.Y., De Wachter, R. & Vaulot, D. (2001). Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature, 409: 607–610.
- Raho, N., Fraga, S., Abad, J.P. & Marín, I. (2018). Biecheleria tirezensis sp. nov. (Dinophyceae, Suessiales), a new halotolerant dinoflagellate species isolated from the athalassohaline Tirez natural pond in Spain. European Journal of Phycology, 53: 99–113.
- Reñé, A., Timoneda, N., Sampedro, N., Alacid, E., Gallisai, R., Gordi, J., Fernandez, A.D., Pernice, M.C., Flo, E. & Garcés, E. (2021). Host preferences of coexisting Perkinsea parasitoids during coastal dinoflagellate blooms. Molecular Ecology. doi: https://doi.org/10.1111/mec.15895.
- Reynolds, E.S. (1963). The use of lead citrate at high pH as an electron opaque stain in electron microscopy. Journal of Cell Biology, 17: 208–212.
- Rhiel, E. (2017). On the extrusomes of Oxyrrhis marina (Dinophyceae). Protoplasma, 254: 901–909.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61: 539–542.
- Siano, R., Kooistra, W.H.C.F., Montresor, M. & Zingone, A. (2009). Unarmoured and thin-walled dinoflagellates from the Gulf of Naples, with the description of Woloszynskia cincta sp. nov. (Dinophyceae, Suessiales). Phycologia, 48: 44–65.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312–1313.
- Takahashi, K., Moestrup, Ø., Jordan, R.W. & Iwataki, M. (2015). Two new freshwater woloszynskioids Asulcocephalium miricentonis gen. et sp. nov. and Leiocephalium pseudosanguineum gen. et sp. nov. (Suessiaceae, Dinophyceae) lacking an apical furrow apparatus. Protist, 166: 638–658.
- Takahashi, K., Moestrup, Ø., Wada, M., Ishimatsu, A., Nguyen, V.N., Fukuyo, Y. & Iwataki, M. (2017). Dactylodinium pterobelotum gen. et sp. nov., a new marine woloszynskioid dinoflagellate positioned between the two families Borghiellaceae and Suessiaceae. Journal of Phycology, 53: 1223–1240.
- Takahashi, K., Sarai, C. & Iwataki, M. (2014). Morphology of two marine woloszynskioid dinoflagellates, Biecheleria brevisulcata sp. nov. and Biecheleriopsis adriatica (Suessiaceae, Dinophyceae), from Japanese coasts. Phycologia, 53: 52–65.
