Abstract
A method to rapidly assess the oligomeric composition of multimeric proteins is notably absent from reported schemes for high throughput production and crystallization of membrane proteins. In this report we have investigated the suitability of PFO-PAGE electrophoresis for this purpose and present examples where it proves highly informative in selecting conditions favouring the functional oligomeric state of the target protein. Features such as the ability to analyze several samples in parallel, including crude membrane extracts, suggest it will be highly adaptable to high throughput analysis of membrane proteins.
| Abbreviations | ||
| PFO | = | perfluorooctanoic acid |
| PAGE | = | polyacrylamide gel electrophoresis |
| SDS | = | sodium dodecylsulphate |
| SEC | = | size exclusion chromatography |
| DDM | = | dodecylmaltoside |
| LDAO | = | lauroyl-N,N-Dimethylamine-N-oxide |
| C12E8 | = | octaethylene glycol monododecyl ether |
| FOS-CHOLINE-16 | = | n-Hexadecylphosphocholine |
| Lyso-FOS-CHOLINE-12 | = | 1-Lauroyl-2-Hydroxy-sn-Glycero-3-Phosphocholine |
| Tris | = | tris(hydroxylmethyl)aminomethane |
| HSV | = | Herpes Simplex Virus |
| CNE | = | clear-native electrophoresis |
Introduction
Membrane proteins serve many crucial functions in the cell, their physiological importance reflected by the fact that over 60% of drugs are believed to act by binding to a cell surface receptor Citation[1]. Therefore it is important for the development of future drugs that structure determination efforts be focussed on membrane proteins.
A significant bottleneck that has hindered crystallization of membrane proteins is their tendency when detergent solubilised to aggregate randomly rather than productively form contacts in a crystal lattice. Protein crystallization has been shown to be favoured under conditions where the protein has a monodisperse oligomeric composition with few large aggregated particles present Citation[2]. Methods that can rapidly assess the oligomeric status of a protein are therefore of great merit to the crystallographer in helping select the best conditions and the best targets for crystallization. However, many of the methods that are routinely applied to soluble proteins such as dynamic light scattering Citation[3] and thermal unfolding (‘the thermofluor assay’ Citation[4]) are complicated to apply to membrane proteins owing to the background signal from detergent micelles. Reports where thermofluor has been applied to membrane proteins are currently limited Citation[5]. It appears that size-exclusion chromatography (SEC) is currently the most widely used alternative technique Citation[6–8].
A drawback of SEC is the amount of time, detergent and protein consumed with the analysis of each sample. Although these limitations have been addressed by a recently described high-throughput ultracentrifugation assay Citation[9], the assay provides limited information as to the exact oligomeric composition of the sample. Such information is important for multimeric membrane proteins, where it is desirable to verify that the native oligomeric state is predominant in a sample.
An alternative method to examine the oligomerization of membrane proteins, PFO-PAGE electrophoresis, has been found to faithfully report the oligomeric state of a number of structurally characterized membrane proteins Citation[10]. Its suitability for the identification of polydisperse samples, which are unlikely to crystallize, remains to be determined. In this report we describe several instances where PFO-PAGE and SEC in fact reach entirely consistent conclusions as to the monodispersity and suitability for crystallization of samples of multimeric membrane proteins. We conclude that PFO-PAGE can thus serve as a valuable alternative to SEC in pre-crystallization screening, with the advantages that it consumes smaller quantities of protein, is able to analyze several samples in parallel and can easily be used to analyze crude membrane extracts.
Materials and methods
Materials
The sodium salt of perfluorooctanic acid (PFO) was from Fluorochem. The New England Biolabs PAGE ruler prestained molecular weight marker was used as a size marker on immunoblots. The BL21(DE3)* and T7 express host strains were obtained from Invitrogen and New England Biolabs respectively. The anti-HSV and anti-T7 antibodies were obtained from Merck Biosciences and an anti-mouse-alkaline phosphate conjugate antibody from Sigma. Immunoblots were developed with solutions of BCIP/NBT (5-Bromo-4-chloro-3-indolyl phosphate/Nitro blue tetrazolium), prepared from SIGMAFAST BCIP/NBT tablets, obtained from Sigma. Acrylamide stock solutions were obtained from BioRad. DDM, FOS-CHOLINE-16 and Lyso-FOS-CHOLINE-12 were from Anatrace; LDAO was from Sigma. The Superdex 200 HR10/300 size exclusion column used for all of the SEC experiments was from GE Healthcare Life Sciences.
PFO-PAGE
The methodology for PFO-PAGE electrophoresis used here closely follows that described by Ramjessingh et al. Citation[10]. Gels were cast using a BioRad Mini Protean apparatus, without any stacking gel, the resolving gel containing 0.375 M Tris pH 8.8 and 0.5% PFO. A 19:1 ration of mono:bis acrylamide was present in the stock solution used for casting. The same running buffer and sample loading buffers were used as described by Ramjessingh et al. Where it was desired to examine denatured samples SDS was added to samples from a 20% w/v stock to a final concentration of 2% and the sample boiled for 1 min. Gels were run in a cold room at no more than 80 V constant voltage. As a precaution gels were carefully monitored during running to check for any cracks developing in the gel.
Isolation of membranes and analysis of detergent extracts
BL21(DE3)* or T7 express were used as host strains for protein expression. Cell membranes were typically isolated from one litre of cell culture by resuspending the cell pellet in approx. 4 ml of buffer per gram of cells containing Roche Complete Protease inhibitor, DNase, RNase and lysozyme (approx. 5 mg per gram of cells). Cells were then lysed with a probe sonicator, the lysate centrifuged at 10,000 g for 20 min and the supernatant then filtered through a 0.45 µm syringe filter. Membranes were then harvested by spinning at 100,000 g for 1 h at 4°C, resuspended in lysis buffer and harvested again with a second 100,000 g spin, followed by rapid freezing in liquid nitrogen and storage at -20°C.
To examine crude detergent extracts by PFO-PAGE, typically approx. 100 µl of membranes were resuspended in 1 ml of buffer and then detergents were added to the desired final concentration to 100 µl aliquots of the membrane suspensions. After centrifugation in a microultracentrifuge at 100,000 g typically 5 µl of supernatant was mixed with 2× sample buffer and loaded on the gel.
Size exclusion chromatography
For all SEC experiments approx. 50 µg of Ni-NTA purified protein in up to 200 µl of buffer was injected onto a Superdex 200 HR10/300 size exclusion column at a flow rate of 0.5 ml/min. The column was equilibrated in the same buffer in which the protein sample was Ni-NTA purified. 0.5 ml fractions were collected.
Results
We have used PFO-PAGE in parallel with analytical size exclusion chromatography to analyze two classes of multimeric membrane transport protein on which the MPSI (membrane protein structure initiative) membrane protein structural genomics consortium has been particularly focussed; firstly, CorA magnesium ion channels and secondly, ABC transporters. We have used PFO-PAGE to analyze both purified proteins and to analyze crude detergent extracts from mixed membranes. For crude extracts immunoblots have been used after electrophoresis to detect the overexpressed membrane protein. For purified proteins, distinct bands were detected when as little as 100 nanograms of protein was loaded on silver stained gels.
To investigate whether aggregated species were detected by this method, samples were analysed in parallel with and without pre-treatment with SDS. The lack of well resolved bands without SDS pre-treatment and a sharp monomer subunit band in the SDS-treated sample would be diagnostic of an aggregated sample. For both CorA and the ABC transporters, we found that boiling with SDS before PFO-PAGE resulted in detection of just the monomeric component subunits (results not shown). Any aggregation can therefore be detected by comparing samples with or without SDS treatment.
CorA ion channels – use of PFO-PAGE to assist choice of detergent for purification
CorA magnesium ion channels are believed to function as either pentamers or tetramers Citation[11], Citation[12]. We have expressed the corA ion channels from two species, the bacterium Pseudomonas Aeruginosa (PA5268) and the thermophilic archeon Methanococcus Jannaschii (MJ1033). After affinity purification in DDM MJ1033 reveals a well resolved band on PFO-PAGE with a reasonable mobility relative to soluble marker proteins to be the anticipated tetramer or pentamer (160–200 kDa) (A). The same sample elutes from a size exclusion column at a reasonable volume to be the tetramer or pentamer (B). On the other hand when the protein is purified in LDAO it appears by PFO-PAGE to be predominantly monomeric. Size exclusion chromatography analysis of the same sample is consistent with the absence of any tetramer or pentamer, the major peak eluting from the column after the 150 kDa marker protein gamma-globulin. PFO-PAGE and SEC analysis therefore consistently conclude that the integrity of the transporter is preserved in DDM but not LDAO. DDM therefore appears to be a safer choice of detergent for purification and crystallization of MJ1033.
Figure 1. Analysis of purified CorA ion channels from M. Jannaschii (MJ1033) and P. Aeruginosa (PA5268) by PFO-PAGE and size exclusion chromatography. (A) Silver stained 7.5% PFO-PAGE gel of purified MJ1033. Whereas DDM-purified MJ1033 shows a predominant band assignable to a tetramer (160 kDa) or pentamer (200 kDa), LDAO-purified MJ1033 appears predominantly monomeric; additional weaker bands may represent a dimer, a tetramer and a pentamer. (B) Superdex 200 size exclusion chromatography of MJ1033 purified in LDAO (light grey dotted line) and DDM (blue solid line) and PA5268 purified in LDAO (black triangles). The position of the void volume of the column (600 kDa) and the elution volume of a 150 kDa marker protein are shown. Consistent with the PFO-PAGE, the major peak at 11 ml in DDM-purified MJ1033 has a retention volume consistent with a tetramer or pentamer, whereas the LDAO-purified MJ1033 has a major peak at 13 ml eluting after the 150 kDa marker; the smaller peak at 11 ml may represent the fainter putative tetramer and pentamer PFO-PAGE bands. (C) Anti-T7 Immunoblot of an 11% PFO-PAGE gel of purified PA5268 and MJ1033, with the position of bands in a prestained molecular weight ladder indicated. MJ1033, but not PA5268, reveals a non-covalent oligomer as a high molecular weight band removed by SDS treatment (marked with an asterisk). Faint high molecular weight bands are observed in the immunoblot of LDAO-purified PA5268; these presumably represent an impurity rather than a non-covalent oligomer as they are observed both with and without SDS-treatment of the sample. This Figure appears in colour in the online version of Molecular Membrane Biology.
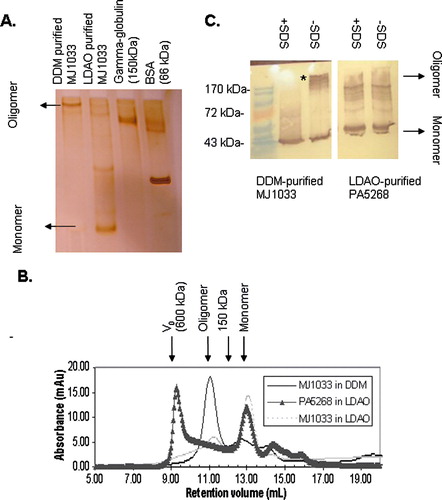
CorA ion channels – PFO-PAGE to assist choice of targets
In contrast to M. Jannaschii CorA, the P. Aeruginosa CorA PA5268 could only be purified successfully in LDAO detergent. The protein could be extracted from membranes with DDM but could not be purified to homogeneity. By PFO-PAGE the LDAO-purified PA5268 shows a predominant band for a monomer (C) and reveals no hint of oligomerization, the sample appearing identical even after boiling it in SDS. By SEC the same sample appears to be aggregation prone as it elutes primarily in the column void volume (B). It is possible the sample may appear monomeric on PFO-PAGE because any weak non-specific interactions between aggregates might be disrupted by PFO-detergent. Regardless, neither a randomly aggregated protein nor isolated monomers represent the oligomeric state of interest and therefore by both techniques MJ1033 appears to be the better candidate for structural studies.
ABC transporters – PFO-PAGE to assist selection of detergent conditions for purification
The majority of ABC transporters used for the import of substrates into bacteria are oligomers assembled from a cytoplasmic ‘ABC’ (ATP Binding Cassette) subunit and an integral membrane spanning subunit Citation[13]. To be amenable to purification the quarternary structure of an overexpressed transporter must be maintained upon extraction from the membrane with detergent. We have found that the integrity of the transporter can be conveniently assessed, prior to undertaking any purification, by parallel analysis of crude detergent extracts using PFO-PAGE.
An example of this is shown in for the P. Aeruginosa molybdenum importer ModBC. Membranes containing overexpressed ModBC were treated with buffers containing different detergents and the integrity of the extracted transporter in each condition analyzed by a PFO-PAGE (A) gel immunoblotted against an HSV epitope on the ABC subunit. A band assignable, in terms of its mobility, to the intact transporter is observable in extracts containing Triton X-100, Zwittergent-3,12, C12E8 and Lyso FOS-CHOLINE 12. Consistent with this, size exclusion chromatography shows the ModBC complex to be intact in C12E8, the integral membrane and ABC subunits of C12E8-purified ModBC co-eluting together at a retention volume consistent with the size of the intact transporter (B). On the other hand, when ModBC is purified in DDM the complex dissociates on size exclusion chromatography, the subunits eluting from the column at markedly different positions. The lower stability of the complex in DDM versus C12E8 is also apparent from PFO-PAGE, the band corresponding to the complex being considerably weaker in the DDM extracts (A), the protein appearing to be predominantly dissociated into its monomeric components. Therefore PFO-PAGE and size-exclusion chromatography analysis consistently identify C12E8 as a better detergent than DDM for preservation of the integrity of the complex.
Figure 2. Selection of detergent for purification and crystallization of ABC transporter ModBC. (A) Anti-HSV immunoblot of a PFO-PAGE analysis of detergent extracts from crude Escherichia coli membranes containing overexpressed ModBC. A band corresponding to the intact ModBC complex is only apparent in Triton X-100, C12E8, Zwittergent-3,12 and Lyso FOS-CHOLINE-12. In some cases there are two oligomeric species identifiable, the faster running band having a reasonable mobility to represent the fully assembled transporter (130 kDa). The upper species (marked with an asterisk) may represent a dimer of fully assembled transporters; the capacity of fully assembled ABC transporters to form strong dimers has been reported elsewhere Citation[15], Citation[16]. (B) Superdex 200 analytical size exclusion chromatogram of ModBC after Ni-NTA purification in C12E8 and DDM. SDS-PAGE shows ModB and ModC subunits co-eluting in the peak in the highlighted portion of the C12E8 SEC chromatogram. In DDM the subunits elute separately in the asterisk-highlighted peaks at 7 ml (ModC) and 13 ml (ModB). This Figure appears in colour in the online version of Molecular Membrane Biology.
![Figure 2. Selection of detergent for purification and crystallization of ABC transporter ModBC. (A) Anti-HSV immunoblot of a PFO-PAGE analysis of detergent extracts from crude Escherichia coli membranes containing overexpressed ModBC. A band corresponding to the intact ModBC complex is only apparent in Triton X-100, C12E8, Zwittergent-3,12 and Lyso FOS-CHOLINE-12. In some cases there are two oligomeric species identifiable, the faster running band having a reasonable mobility to represent the fully assembled transporter (130 kDa). The upper species (marked with an asterisk) may represent a dimer of fully assembled transporters; the capacity of fully assembled ABC transporters to form strong dimers has been reported elsewhere Citation[15], Citation[16]. (B) Superdex 200 analytical size exclusion chromatogram of ModBC after Ni-NTA purification in C12E8 and DDM. SDS-PAGE shows ModB and ModC subunits co-eluting in the peak in the highlighted portion of the C12E8 SEC chromatogram. In DDM the subunits elute separately in the asterisk-highlighted peaks at 7 ml (ModC) and 13 ml (ModB). This Figure appears in colour in the online version of Molecular Membrane Biology.](/cms/asset/34c4ed96-3b6e-4bce-a737-9fc024eaa858/imbc_a_345021_f0002_b.jpg)
Discussion and conclusion
When applied to CorA ion channels and ABC transporters PFO-PAGE has been found to be a valuable qualitative method to probe whether a multimeric target protein is in its functional oligomeric state. For these two classes of protein PFO-PAGE has been found to reliably inform the selection of both the target and the buffer detergent conditions for structural studies. The only instance of a discrepancy between SEC and PFO-PAGE analysis was for the LDAO-purified CorA ion channel PA5268, which appears monomeric by PFO-PAGE but randomly aggregated by SEC analysis. This indicates PFO may disrupt the weak interactions between subunits in a non-specifically aggregated protein sample, making it appear to be monomeric on PFO-PAGE. PFO-PAGE might therefore not be the best method for the analysis of proteins whose functional state is monomeric, as it may not be able to distinguish a polydisperse aggregated sample from a monodisperse monomeric sample better suited for crystallization. For the analysis of multimeric proteins, on the other hand, it is typically not important to distinguish monomers from random aggregates, neither representing the functional oligomer of interest.
Although PFO may disrupt non-specific aggregates, the results presented here and by Ramjessingh et al. Citation[10] show PFO-PAGE does not affect the specific interactions between the subunits in a macromolecular complex. Furthermore, our results show PFO-PAGE provides a sensitive and valuable monitor of any effect of the purification detergent on such interactions. This may set it apart from high resolution clear-native electrophoresis Citation[14] (CNE) in which a mixture of deoxycholate with either DDM or Triton X-100 is used in the running buffer. The oligomeric composition reported by CNE has been found to be dependent on the detergent composition chosen for the electrophoresis buffer.
Overall, we have found PFO-PAGE to not only be highly informative but also to be convenient to implement in a number of respects. Firstly, it consumes only nanogram quantities of protein per sample, which is considerably less than required for a typical size exclusion chromatography experiment. Secondly, it can be used to analyze many samples in parallel and is easily used in conjunction with immunoblotting to analyze partially purified samples. Finally, the method is relatively undemanding technically and does not require expensive instrumentation. It is conceivable that PFO-PAGE could be routinely incorporated as a qualitative assay into a scheme for high throughput purification and crystallization of membrane protein targets.
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council (grant number BBS/B/14418), and is part of the MPSI membrane protein structural initiative. We thank the University of Manchester, Faculty of Life Sciences for supporting studentships (to M.S. and J.K.). We thank Dr Mark Young for valuable advice on the conditions for PFO-PAGE electrophoresis. We thank Dr Peter Roach and Professor Peter Henderson at the University of Leeds for growing fermenter cultures overexpressing ModBC. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nature Rev Drug Discov 2001; 1: 198–210
- D'Arcy A. Crystallizing proteins – a rational approach”. Acta Crystallographica 1994; D50: 469–471
- Ferré-D'Amaré AR, Burley SK. Dynamic light scattering in evaluating crystallizability of macromolecules. Meth Enzymol 1997; 276: 157–166
- Geerlof A, Brown J, Coutard B, Egloff M, Enguita F, Fogg M, Gilbert R, Groves M, Haouz A, Nettleship J, Nordlund P, Owens R, Ruff M, Sainsbury S, Svergun D, Wilmanns M. The impact of protein characterization in structural proteomics. Acta Crystallographica 2006; D62: 1125–1136
- Yeh AP, Macmillan A, Stowell MHB. Rapid and simple protein-stability screens: application to membrane proteins. Acta Crystallographica 2006; D62: 451–457
- Lemieux MJ, Song J, Kim MJ, Huang Y, Villa A, Auer M, Li X, Wang D. Three-dimensional crystallization of the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Protein Sci 2003; 12: 2748–2756
- Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 2006; 14: 673–681
- Eshaghi S, Hedren M, Ignatushchenko M, Nasser A, Hammarberg T, Thornell A, Nordlund P. An efficient strategy for high-throughput expression screening of recombinant integral membrane proteins. Protein Sci 2005; 14: 676–683
- Gutmann DA, Mizohata E, Newstead S, Ferrandon S, Postis V, Xia X, Henderson PJ, Veen HWv, Byrne B. A high-throughput method for membrane protein solubility screening: the ultracentrifugation dispersity sedimentation assay. Protein Sci 2007; 16: 1422–1427
- Ramjessingh M, Huan L, Garami E, Bear C. Novel method for evaluation of the oligomeric structure of membrane proteins. Biochem J 1999; 342: 119–123
- Maguire ME. The structure of CorA: a Mg2 + -selective channel. Curr Opin Structural Biol 2006; 16: 432–438
- Warren MA, Kucharski LM, Veenstra A, Shi L, Grulich PF, Maguire ME. The CorA Mg transporter is a homotetramer. J Bacteriol 2004; 186: 4605–4612
- Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem 2004; 63: 241–268
- Wittig I, Karas M, Schägger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Molec Cellular Proteomics 2007; 6: 1215–1225
- Ferreira-PereiraDagger A, Marco S, Decottignies A, Nader J, Goffeau A, Rigaud J. Three-dimensional reconstruction of the Saccharomyces cerevisiae multidrug resistance protein Pdr5p. J Biological Chem 2003; 278: 11995–11999
- Chami M, Steinfels E, Orelle C, Jault J, Pietro AD, Rigaud J, Marco S. Three-dimensional structure by cryo-electron microscopy of YvcC, an homodimeric ATP-binding cassette transporter from Bacillus subtilis. J Molec Biol 2002; 315: 1075–1085