Abstract
Since mutations on POU1F1 gene possibly resulted in deficiency of growth hormone (GH), prolactin (PRL), thyroid-stimulating hormone (TSH) and pituitary specific transcription factor-1 (POU1F1), this study revealed the polymorphism of cattle POU1F1-HinfI locus and analyzed the distribution of alleles on eight cattle breeds including five indigenous Chinese cattle and three crossbred cattle breeds. The PCR-RFLP analysis showed that allele B was the dominant allele. The frequencies of allele B varied from 0.535 to 0.868. Further study, distributions of genotypic and allelic frequencies at this locus were found to be significantly different among populations based on a χ 2-test (p<0.05), suggesting that the breed factor significantly affected the molecular genetic character of POU1F1 gene. The genetic diversity analysis revealed that these populations had a wide spectrum of genetic diversity in cattle POU1F1-HinfI locus. In addition, the relationship between the polymorphism of POU1F1 gene and milk production traits in Chinese Holsteins was also analyzed. The effects of POU1F1 gene on milk yield, fat percent, pre somatic cell count, somatic cell count and somatic cell score were not homogeneous(p>0.05). And, POU1F1 gene had homogeneous effect on protein percent (p<0.05). Protein percent of individuals with genotype AB was higher than those with BB.
Abbreviations used: POU1F1, pituitary specific transcription factor-1; PCR, Polymerase Chain Reaction; RFLP, Restriction Fragment Length Polymorphism; GH, growth hormone; PRL, prolactin; TSH, thyroid-stimulating hormone; QQ, Qinchuan cattle; HC, Chinese Holstein cattle; XB, Xinjiang Brown cattle; NY, Nanyang cattle; JX, Jiaxian Red cattle; AQ, crossbred cattle from Angus and Qinchuan Cattle; DQ, crossbred cattle from German yellow cattle and Qinchuan Cattle; LQ, crossbred cattle from Limousin and Qinchuan cattle; PIC, polymorphism information content; LSM, Least square means; SE, standard error
POU1F1 (also named PIT-1) is a positive regulator for growth hormone (GH), prolactin (PRL) and thyroid-stimulating hormone (TSH) and itself in mammalian animals (Cohen et al. Citation1997). Accordingly, mutations on this gene possibly result in deficiency of GH, PRL, TSH and POU1F1 (Cohen et al. Citation1997, Li et al. Citation1990). It had been reported that mutations within GH, PRL, TSH genes were significantly associated with growth, development, lactation and cashmere traits in mammalian animals, therefore, mutations of POU1F1 gene were shown to associate with mice Snell dwarf (dw), mice Jackson dwarf (dw-J), human dwarfism (Li et al. Citation1990 and Pfaffle et al. Citation1992), swine growth and meat performance (Stancekov et al. Citation1999), bovine growth (Zhao et al. Citation2004). Therefore, the POU1F1 gene was suggested to be the potential candidate gene for studying production traits in domestic animals.
Qinchuan cattle (QQ) are one of five Chinese native yellow cattle breeds. It had been classified as national resource to be protected because of its valuable. It has tall physique,tender meat quality and clear marbling. Meat-bone ratio, lean meat percentage and eye muscle area of the cattle are close to or higher than those of foreign beef breeds. Chinese Holsteins (HC) came from purely breeding of Holsteins which were introduced from America, Canada, Japan, Australia, New Zealand, Russia, Germany, and Italy and so on. And they also came from advanced-generation upgrading of those Holsteins with native yellow cattle. Chinese Holsteins which have well-proportioned frame and obvious milk trait are excellent milk breeds.
To date, few polymorphism of POU1F1 gene within Qinchuan cattle and Chinese Holsteins had been reported. The objective of this study was to detect the HinfI polymorphisms within POU1F1 gene in cattle, which possibly contributed to conducting association analysis and evaluating it as genetic marker in production traits for breeding and genetics of meat and milk cattle. Moreover, considering the important effect on growth development and the positive regulations of GH, PRL and TSH gene, the relationship between the polymorphism of milk production traits in Chinese Holsteins was also analyzed.
Materials and Methods
DNA Samples
Genomic DNA samples were obtained from 218 unrelated cattle belonging to two breeds: Chinese Holstein cattle (HC) and Qinchuan cattle (QQ) reared in the province of Jiangsu and Shaanxi (China). DNA samples were extracted from leucocytes according to (Mullenbach et al. Citation1989).
PCR conditions
According to the report from (Renaville et al. Citation1997), a pair of primers (F: 5′–AAA CCA TCA TCT CCC TTC TT -3′and R: 5′–AAT GTA CAA TGT GCC TTC TGA G -3′) were synthesized to amplify a 451 bp product from 5 intron and 6 exon. The PCR was performed in a 25 µl reaction mixture containing 10 pmol of each primer, 200 µM dNTP (dATP, dTTP, dCTP and dGTP), 1×buffer (including 1.5 mM MgCl2), 1 units Taq DNA polymerase and 100 ng cattle genomic DNA as template. The amplification was carried out for 35 cycles at the conventional PCR protocol with annealing at 53.5°c.
Genotyping of HinfI POU1F1 allele by PCR-RFLP
Aliquots of 10 µl PCR products of POU1F1 gene were digested with 8U HinfI (MBI fermentas) for 5h at 37°c following the supplier's directions. The digested products were detected by electrophoresis in 3.0% agarose gel stained with ethidium bromide.
Statistical analysis
Genotypic frequencies, allelic frequencies and Hardy–Weinberg equilibriums were directly calculated. Differences for these frequencies at gPOU1F1-HinfI locus between populations were analyzed using a χ 2-test, which were performed by SPSS software (version13.0). Population genetic indexes, namely, gene homozygosity, gene heterozygosity, effective allele numbers, and PIC (Polymorphism Information Content), were calculated by Nei methods (Nei and Roychoudhurg et al. Citation1974, Nei and Li et al. Citation1979) not only involving the two analyzed breeds, but also the six reported breeds such as Xinjiang Brown cattle (XB), Nanyang cattle (NY), Jiaxian Red cattle (JX), crossbred cattle from Angus and Qinchuan Cattle (AQ), crossbred cattle from German yellow cattle and Qinchuan Cattle (DQ) and crossbred cattle from Limousin and Qinchuan cattle (LQ). The statistical software SPSS (Version 13.0) was also used to analyze the relationship between the genotypes and milk production traits in Chinese Holsteins. The adjusted Linear Model with fixed effects was used to deal with the relationship between genotypes and milk production traits of Chinese Holstein cattle. Linear Model: Y ijk = µ + G i +S j +L k + ϵ ijk , where Y ijk was the trait measured on each of the ijkth animal, µ was the overall mean, G i was the type of the ith genotype, S j was the type of the jth sex; L k was the type of the kth parity and ϵ ijk was the random error.
Results and Discussion
The polymorphism of POU1F1 gene in 8 cattle breeds
The 451 bp products () were amplified using the specific combination of primers and the digestion with HinfI endonuclease showed one fragment (451 bp) for A allele and two fragments (244 and 207 bp) for B allele. Therefore, genotype AA and BB demonstrated one and two bands, respectively. While genotype AB showed three bands (). In this study, the frequencies of AA/AB/BB for QQ and HC were 0.030/0.403/0.567 and 0.007/0.252/0.742, respectively. Accordingly, their frequencies of allele B were 0.769 and 0.868, respectively. Based on the genotypes of the six cattle breeds previously reported (Lin et al. Citation2009, Xue et al. Citation2006, Qiu et al. Citation2009, Liu et al. Citation2005) and the present genotypic frequencies, the distributions of genotypic and allelic frequencies of gPOU1F1-HinfI locus in 8 breeds were shown ().
Figure 1. The PCR products of POU1F1 gene. Lanes 1-10 = PCR amplification products of POU1F1 gene (451bp); Lane 11= maker (1200, 900, 700, 500, 300, and 100bp).
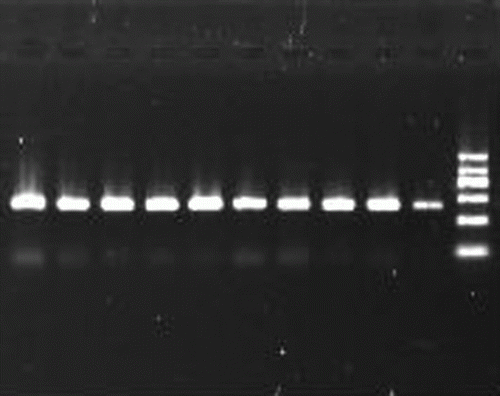
Figure 2. HinfI digestion patterns of PCR products. Lane 2= genotype AA(451bp);Lanes 3,7,9 = genotype AB(451, 244 and 207bp); Lanes 4,5,6, 8, 10 = genotype BB(244 and 207bp);Lane 1= marker (1200, 900, 700, 500, 300, and 100bp).
Figure 3. The distribution of genotypic (a) and allelic frequencies (b), genetic diversity (c) of POU1F1-HinfI locus in 8 breeds. Note: The genotypic and allelic frequencies of XB, NY, JX, AQ, DQ, LQ, were cited from reference (Lin et al. Citation2009, Xue et al. Citation2006, Qiu et al. Citation2009 and Liu et al. Citation2005).
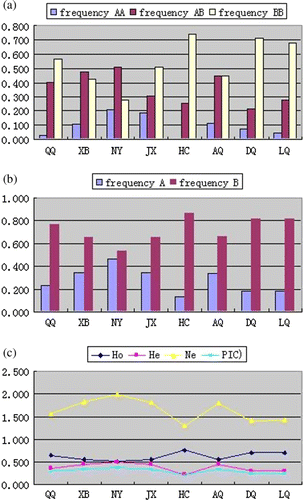
Genotype BB was predominant in QQ, JX, HC, DQ and LQ while genotype AB was predominant in XB and NY. However, genotype AB and BB were equal in AQ while genotype AA was fewer. Genotypic frequencies for the various polymorphisms at POU1F1-HinfI locus were found to be significantly different among the eight populations based on a χ 2-test (χ 2=90.114, df =14, P<0.001). Significant differences for allelic frequencies among the populations were also revealed (χ 2=83.422, df=7, P<0.001). Moreover, there were significant differences for genotypic and allelic frequencies between QQ, XB, NY, JX, HC, AQ, DQ and LQ populations (P<0.05) (). So, there were significant differences for genotypic and allelic frequencies at cattle POU1F1-HinfI locus in the eight cattle breeds. It was obvious that the breed factor significantly affected the distribution of genotypic and allelic frequencies at cattle POU1F1-HinfI locus.
Table 1. χ 2 and P values differences for genotypic and allelic frequencies among 8 cattle breeds at POU1F1-HinfI locus.
In present populations, gene homozygosty, gene heterozygosty, Effective allele numbers and polymorphism information content (PIC) were shown in () and (). Comparisons of genetic diversity of above cattle breeds demonstrated that HC had the highest homozygosty and lowest PIC, while NY had the lowest homozygosty and highest PIC. The result indicated that HC population was in a good homologous status, while NY cattle breed was not. This may due to breeding. Chinese Holstein cattle were selected largely in the long-term artificial selection process, which tended to be homozygous and then fixed gradually, thus appeared the conservatism of breed. With the vast spectrum of variation, NY cattle breed had a wider breeding space. According to the classification of PIC (low polymorphism if PIC value < 0.25, median polymorphism if 0.25 < PIC value < 0.5, and high polymorphism if PIC value > 0.5) (Vaiman et al. Citation1994), only HC possessed poor genetic diversity, while the others belonged to middle genetic diversity. In addition, we found that the crossbred cattle populations still had abundant genetic diversity. It suggested that we could increase the intensity of artificial selection to get the superior breeds. At this locus, all the population was at Hardy-Weinberg equilibrium except the JX cattle population. We take it for granted that this was also due to the selection.
Table 2. Genetic diversity indices at POU1F1-HinfI locus in 8 populations.
The relationship between the polymorphism of POU1F1 gene and milk production traits in Chinese Holsteins
The relationship between the polymorphism of POU1F1 gene and milk production traits in Chinese Holsteins was also analyzed. Least square means and standard errors for protein percent of various POU1F1 genotypes were presented in (). Significant statistical differences were found in protein percent. Multiple comparisons results indicated that individuals with genotype AB were significantly higher than those of individuals with genotype BB in protein percent (P < 0.05) (). It suggested that allele A had a positive effect on protein percent, which was similar to the study of Renaville's (Renaville et al. Citation1997). So, we can take POU1F1 gene as a reliable molecular marker of the protein percent selection. However, no statistical differences were found in the other milk composition (P > 0.05) (). This result was different with that from Renaville (Renaville et al. Citation1997). Further investigations must be essential for detecting the polymorphism of this gene in a lager and broader variety of cattle breeds and populations.
Table 3. Least square means and standard errors for protein percent of various POU1F1 genotypes.
Table 4. F value for significance of the sources of variation for effect analysis of the other milk composition under least square model.
Conclusion
We detected the HinfI polymorphisms within POU1F1 gene in 8 cattle breeds in this study. Genotypic frequencies, allelic frequencies and Hardy–Weinberg equilibriums were directly calculated. And, genetic diversity indices at POU1F1-HinfI locus were also presented. There were significant differences for genotypic and allelic frequencies at cattle POU1F1-HinfI locus in the eight cattle breeds. It was obvious that the breed factor significantly affected the distribution of genotypic and allelic frequencies at cattle POU1F1-HinfI locus. In addition, association analysis between POU1F1 gene polymorphism and six milk production traits of Chinese Holstein cattle samples was performed, and the results indicated that the allele A had a positive effect on protein percent. It can be taken as a reliable molecular marker of the protein percent selection. However, the relationships between genotypes and the other milk production traits including milk production had not been found.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 30972080, 30901023), National Key Technology R&D Program (Grant No. 2008ADB2B03-19), Keystone Project of Transfergene in China (Grant No. 2009ZX08009-157B, 2008ZX08007-002, and 2009ZX08007-005B-07), Program of National Beef Cattle Industrial Technology System (Grant No. CARS-38) and Science and Technology Industrialization (Agriculture) Program of Nantong (No. CL2009002).
Additional information
Notes on contributors
X.T. Fang
L.J. Yan and X.T. Fang contributed equally to this workReferences
- Cohen , LE , Wondisford , FE and Radovick , S . 1997 . Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin . Endocrinology Metabolism Clinics of North America , 25 : 523 – 540 .
- Li , S , Crenshaw , EB , Rawson , EJ , Simmons , DM , Swanson , LW and Rosenfeld , M.G . 1990 . Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene Pit-1 . Nature , 347 : 528 – 533 .
- Lin , JP , He , SG , Bai , J , Liu , L , Yang , B , Tan , LX and Liu , MJ . 2009 . Correlations between POU1F1 polymorphism and milk yield in Xinjiang Brown cattle and Chinese Holstein cattle . China Herbivores , 29 ( 1 ) : 6 – 7 .
- Liu , B , Chen , H , Lan , XY , Lei , CZ , Zhang , ZQ and Zhang , RF . 2005 . Correlation of polymorphisms of POU1F1 gene and growth traits in Qinchuan cattle and its hybrid cattle . Scientia Agricultura Sinica , 38 ( 12 ) : 2520 – 2525 .
- Mullenbach , R , Lagoda , PJ and Welter , C . 1989 . An efficient salt-chloroform extraction of DNA from blood and tissue . Trends in Genetics , 5 : 391
- Nei , M and Li , WH . 1979 . Mathematic model for studying genetic variation in terms of restriction endonucleases . Proceedings of the National Academy of Sciences , 76 : 5269 – 5273 .
- Nei , M and Roychoudhurg , AK . 1974 . Sampling variance of heterozygosity and genetic distance . Genetics , 76 : 379 – 390 .
- Pfaffle , RW , DiMattia , GE , Parks , J , Brown , M , Wit , JM , Jansen , M , Vander , NH , Van den Brande , JL , Rosenfel , MG and Ingraham , HA . 1992 . Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia . Science , 257 : 1118 – 1121 .
- Qiu , CY , Chen , H , Pan , CY , Wang , J and Niu , H . 2009 . Genetic variations of HinfI, AluI and PstI loci within POU1F1 gene exon 6 and its association with growth traits in Jiaxian Red cattle . Journal of Northwest A &F University , 37 ( 5 ) : 43 – 48 .
- Renaville , R , Gengler , N , Vrech , E , Prandi , A , Massart , S , Corradini , C , Bertozzi , C , Mortiaux , F , Burny , A and Portetelle , D . 1997 . Pit-1 gene polymorphism, milk yield and conformation traits for Italian Holstein Friesian bull . Journal of Dairy Science , 80 : 3431 – 3438 .
- Stancekov , K , Vasicek , D , Peskovicov , D , Bull , J and Kubek , A . 1999 . Effect of genetic variability of the porcine pituitary-specific transcription factor (PIT-1) on carcass traits in pigs . Animal Genetics , 30 : 313 – 315 .
- Vaiman D , Mecier D , Moazami-Gou darzik Eggena Ciampolini R Ciampolini R , Lepingle A , Velamala R , Kaukinen J , Varvios L , Martin P , Leveziel H , Guerin G. 1994 . A set of 99 cattle microsatellites characterization synteny mapping and polymorphism . Mammalian Genome 5 : 288 – 297 .
- Xue , K , Chen , H , Wang , S , Cai , X , Liu , B , Zhang , F , Lei , CZ , Wang , XZ , Wang , YM and Niu , H . 2006 . Effect of genetic variations of POU1F1 gene on growth traits of Nayang cattle . Journal of Genetics and Genomics , 33 ( 10 ) : 901 – 907 .
- Zhao , Q , Davis , ME and Hines , HC . 2004 . Associations of polymorphisms in the Pit-1 gene with growth and carcass traits in Angus beef cattle . Journal of Animal Science , 82 ( 8 ) : 2229 – 2233 .